AFG3L2 Cure Odyssey
Rita and Daniel launched a yeast-powered drug repurposing project on behalf of their son Milo, who has two pathogenic variants in his AFG3L2 gene. The TargetMol screen is happening this week.
In collaboration with
Six months ago, our friends at Rare Village introduced us to a pioneer family based in the Bay Area. Milo’s parents, Rita and Daniel, are not professional scientists, a trait they share with the vast majority of Perlara’s partner pioneers. Despite that lack of degree or real-world experience in research, they have been up to the challenge. Resilience in the face of an uncertain cure odyssey is a diagnostic biomarker of a biomedical pioneer family.
As shown below in the excerpted whole exome sequencing report, Milo is compound heterozygous for two variants in the AFG3L2 gene. Homozygous loss-of-function AFG3L2 mutations are associated with spastic ataxia and optic atrophy. Heterozygous (autosomal dominant) toxic gain-of-function variants are also associated with disease, consistent with a Goldilocks set point of AFG3L2 activity that must be maintained.
AFG3L2 is a component of the mitochondria’s internal protein shredder system aka a metalloprotease. AFG3L2 is encoded by the yeast ortholog AFG3. Yeast lacking AFG3 are unable to grow in conditions that require functional mitochondria, e.g., media containing lactate as the sole carbon source. Both of Milo’s variants fall in the proteolytic domain which is responsible for chewing up misfolded proteins and unused electron transport chain subunits, either of which run amok if left unchecked.
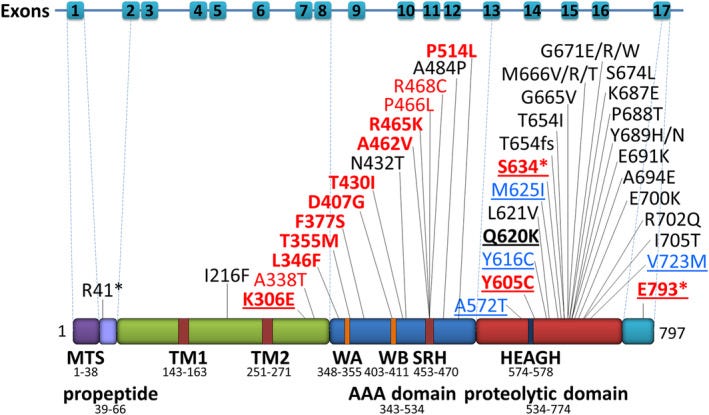
The W583STOP variant is clearly loss-of-function, but his M625I variant — previously not seen before in the published literature — was annotated as a VUS: variant of unknown significance. AlphaFold renders the following predicted 3D structure of the nearly 800-amino-acid AFG3L2 protein, with M625 highlighted in green buried in the heart of the proteolytic domain, which is comprised of a knot of blue alpha helices. Zooming into M625 on the top of two panels on the left, one can appreciate that the methionine side chain inserts in to the space between three cross-crossing alpha helices.
When that intra-helical space is filled in with the side chains of the neighboring amino acids, the yellow sulfur atom of M625 is enmeshed in a hydrogen-bond love triangle with S656 and Q660, which are both hydrogen bonded to each other.
When that intra-helical space is filled in with the side chains of the neighboring amino acids, the sulfur atom of M625 is enmeshed in a hydrogen-bond love triangle with S656 and Q660, which are hydrogen bonded to each other. M625I means the the sulfur-containing thioester side chain of methionine has been replaced by the alkyl side chain of isoleucine, and as a consequence the hydrogen bond network supported by the methionine side chain falls apart. Given that both of Milo’s variants can be assumed to be loss-of-function, we assumed that an AFG3 whole-gene knockout yeast strain lacking any AFG3 protein expression would be the simplest path to a drug repurposing screen. The afg3∆ whole-gene knockout yeast would also afford the opportunity to identify AFG3L2 bypass suppressors, i.e., rescue mechanisms that don’t depend on potentiating residual AFG3L2 activity.
We encountered unexpected turbulence in all but one yeast avatar generation project, to date. For mitochondrial diseases like SURF1, which has an established pattern of homozygous recessive inheritance, instead of generating patient avatars with specific loss-of-function missense or nonsense variants, we reasoned that an off-the-shelf whole-gene deletion mutant was sufficient and stood in as the most representative AFG3L2 yeast avatar. The other advantages of off-the-shelf whole-gene deletion strains are cost and delivery time. We still apply a few QC tests upon arrival to make sure the package label matches its contents. But then its off the races.
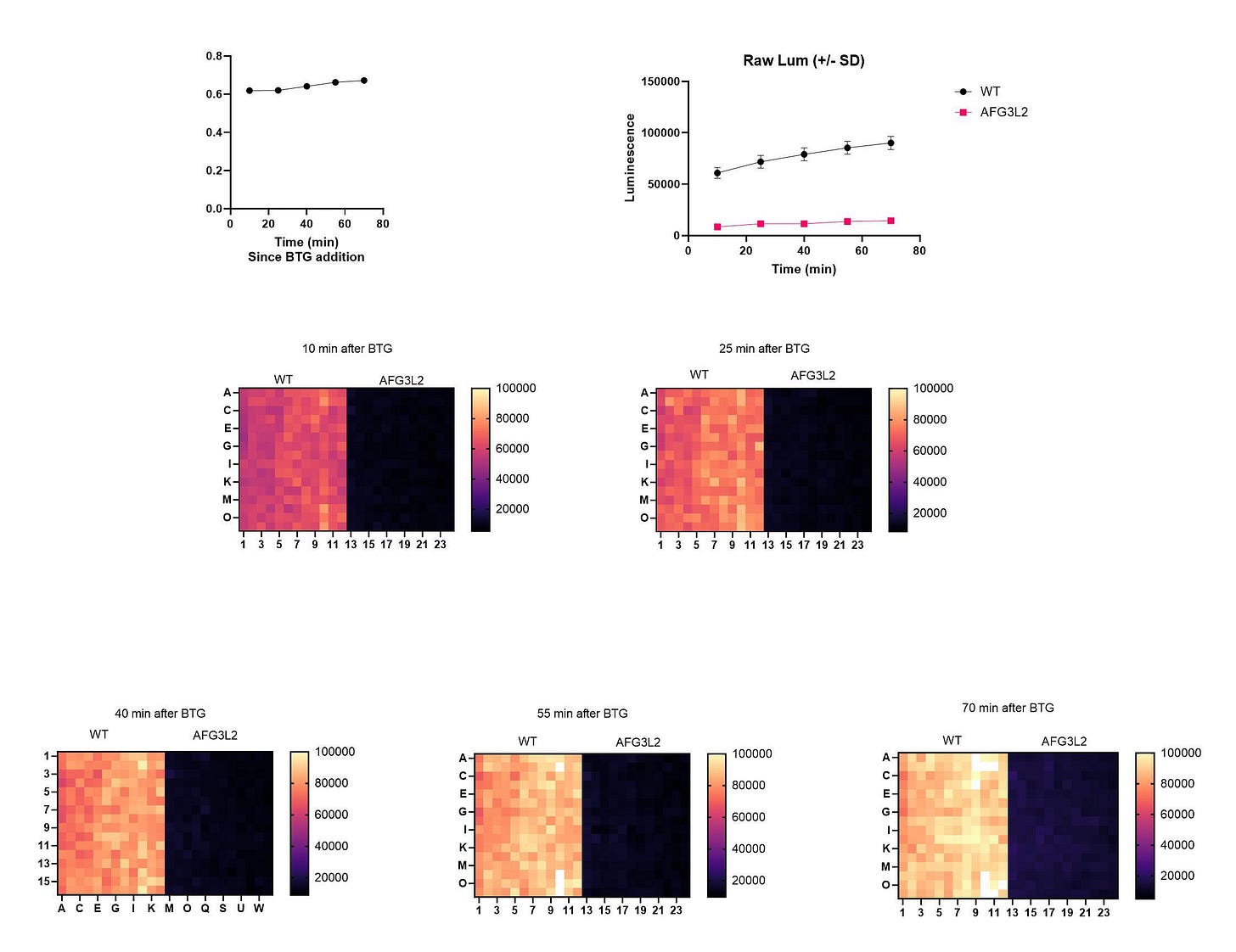
The seeding density experiment went off without a hitch in the first round. We included SURF1 as a positive control. The AFG3L2 yeast knockout strain (afg3∆) and the SURF1 yeast knockout strain (shy1∆) are both unable to grow in media containing 2% lactate as the sole carbon source.
However, the Z’ optimization experiment did not go quite according to plan, in large part because the wildtype control under-performed, i.e., did grow as robustly as expected. Also the afg3∆ mutant’s growth started to creep up at the later timepoints. There can be drift in knockout mutants that are passaged even over a brief period of time in the lab, so we did some troubleshooting. That set us back by a few weeks.
With yeast it’s possible to hit Alt-Ctl-Del by going back to the frozen stock and re-streaking for fresh colonies. In April, we relocated popup lab operations from SFBL to Bad Ass Labs (BAL) in Alameda, temporarily pausing projects for several weeks. We also booted up a collaboration with HTSF at University of California Berkeley to perform drug repurposing screens on the 8,400-compound TargetMol library.

It’s never easy having to explain to a family why timelines are taking longer than expected. It’s actually much harder (and more stressful) to communicate a failed experiment requiring repetition rather than a definitively negative result, which at least closes a door shut rather than leaving it frustratingly ajar. The task of communicating the ups and downs of the research process falls to project leads.
Cure Guide Uche Medoh from Stanford has been up to the challenge. We have these troubleshooting lab meeting style calls with clients all the time to make sure they understand the vicissitudes of lab life. Even the single-cell yeast poses challenges. (So you can just imagine prima donna models like iPSCs).
We repeated the Z’ optimization experiment at BAL and to everyone’s relief got the expected night-and-day contrast between wildtype and the afg3∆ mutant. And there’s no creeping growth by the afg3∆ mutant, indicating no drift in the active population maintained at BAL.
Out of an abundance of caution we had HTSF repeat the Z’ optimization experiment. We got identical if not slightly statistically better results. Rita and Daniel have been patiently waiting for these results which we initially thought would take less than three months. Even a simple model system like yeast can still be an untamed frontier.
After plugging away at this for the past year — failing, learning and iterating as quickly and responsibly as we can — we feel confident that we’ve productized the yeast-powered drug repurposing service. We hope this is the last of the Perlara lab projects that take longer than expected.
If all goes according to plan, we’ll receive results from HTSF just before EOB on Friday July 7, and it will have been worth the wait. Stay tuned!