ALG11-CDG Cure Odyssey
Two yeast drug repurposing projects launched at UCSF in between Perlara 1.0 and 2.0. We're finally ready to run the first drug repurposing screen for ALG11-CDG using an ALG11 knockout yeast avatar.
In collaboration with
Readers of Cure Odysseys know that our yeast-powered drug repurposing service is a good fit for many CDGs. For the newly subscribed, CDG stands for congenital disorder of glycosylation. At present, we are working with nine CDG communities. With 170 known CDG genes and counting, there is much more work to be done.
Shannon and Chase Willardson are the parents of Charlotte and Ava, two girls living with ALG11-CDG. One of the girls was part of a tiny n-of-2 clinical cohort published in a 2019 AJMD paper. Both girls are compound heterozygous for two ALG11 pathogenic variants: E312K and M408R. At the time of that publication, only 10 other ALG11-CDG patients had been described in the medical literature.
Today that number is not much higher, making ALG11-CDG typical of CDGs, and rare diseases generally: more than 10 patients but fewer than 100 identified in the world. What we’ve gotten accustomed to calling rare is actually ultra-rare.
The rules of rare break down for ultra-rares in the same way that the rules of Newtonian physics break down at the scale of subatomic particles. That’s why we need a new generalized framework for rare genetic diseases — no matter how small or big — along the lines of what we’ve been calling 1-to-N medicine.
Not long after that 2019 ALG11-CDG clinical report was published, and a month before the COVID lockdowns began, CDG pioneer families, researchers and clinicians from across “Glycoland” convened at the Dana Point Hotel in San Diego for the 2020 Sanford Burnham Prebys Rare Disease Day Symposium and CDG Family Conference. The air was filled with hope, and we would soon find out, a virus that put everyone’s lives, and the scientific hopes of all rare genetic disease communities, on indefinite hold.
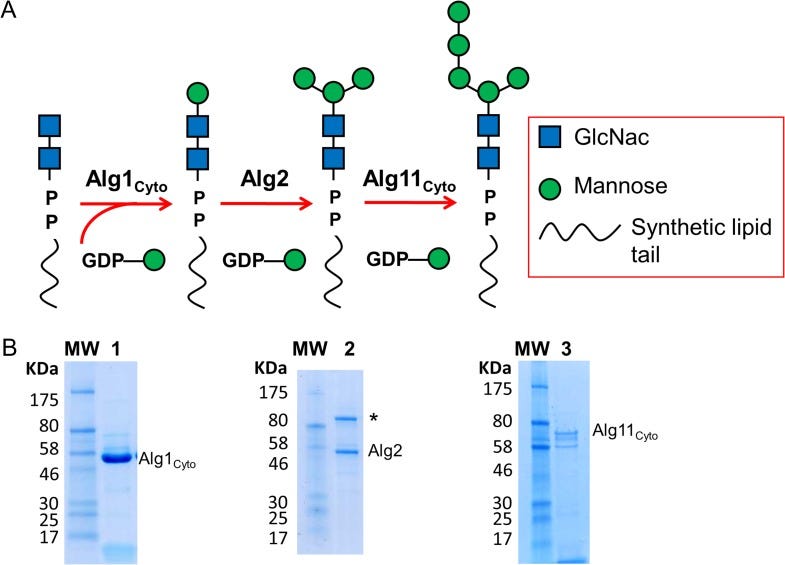
Shannon emailed during the conference to make sure we followed up on the potential for ALG11-CDG yeast avatars after we got home from the meeting. Fresh from our first PMM2-CDG drug repurposing experience that led to the discovery of epalrestat, ALG11 seemed like a perfect fit on paper. ALG11 encodes an enzyme called a mannosyltransferase, which is part of lipid-linked oligosaccharide (LLO) assembly line and forms a complex with two other mannosyltransferases and CDG genes, ALG1 and ALG2.

Zooming in on ALG11, we see that it extends one the glycan’s antennae, or branches. ALG11 is fueled by GDP-mannose, which itself is generated from mannose-1-phosphate by Gemini enzymes GMPPA and GMPPB. Loss of GMPPA or GMPPB each causes a CDG. Mannose-1-phosphate is generated from mannose-6-phosphate by the action of PMM2, which of course causes the most common type of CDG.
If you’re noticing an “all-in-the-family” CDG theme emerge, good. It means you’ve been paying attention.
In the MEPAN yeast drug repurposing project that started side-by-side with ALG11-CDG until the ALG11-CDG project got stuck in troubleshooting mud, Kelly Pan and Dr Tomas Wald were able to create MEPAN patient avatars and demonstrate a gradient of growth defects that were benchmarked to a whole-gene knockout.
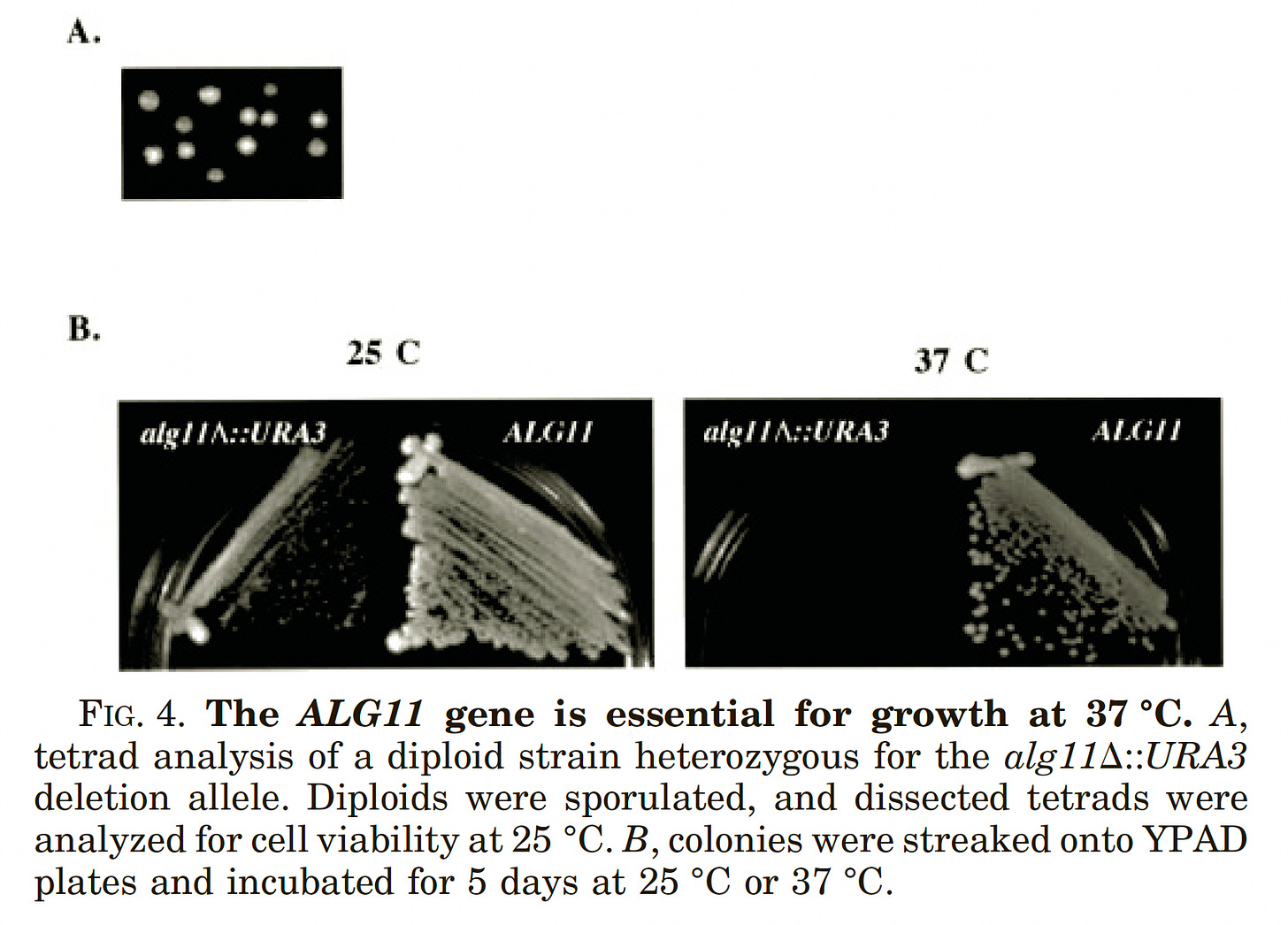
After multiple attempts and restarts, Kelly finally got everything to work in recent weeks. In spotting assays that were published in a paper in 2001, we knew that the ALG11 knockout (KO) yeast avatar has a severe heat-sensitive growth defect when grown at 37˚C. The tetrad analysis (inset A, below) proves that ALG11 is required for cell growth at 37˚C since only two of four spores were viable. The ALG11 KO yeast avatar is also slow-growing at 25˚C.
In contrast to the MEPAN project, we expressed the human ALG11 gene in the ALG11 KO yeast avatar instead of the yeast ALG11 gene. Another thing to keep in mind is that the SEC53 promoter, not the native ALG11 promoter, is driving expression in the ALG11-CDG avatars.
In each of the spotting assay experiments shown below, the ALG11 KO yeast avatar (one row up from the bottom row) behaves as expected and is included as a negative control. The top four rows are independent clones that received the wildtype ALG11 gene. We tested three independent clones of each of the avatars. W303 serves as a wildtype positive control.
Here are the results of the ALG11 E312G yeast avatar at 30˚C and 37˚C. The data show that the ALG11 E312G yeast avatar grows as well as ALG11 wildtype avatar under the experimental conditions that we tested.
Here are the results of the ALG11 M408R yeast avatar at 30˚C and 37˚C. The data also show that the ALG11 M408R yeast avatar grows as well as the ALG11 wildtype avatars under the experimental conditions that we tested.
These results are surprising because we expected at least one of the avatars to have a growth defect based on what we know about how the ALG11 protein functions. Fortuitously, a paper from 2010 almost made the E312G and M408R avatars for us.
Examining E312G first (yellow arrow), it’s next-door neighbors with K313, which is critical for binding activated mannose residues. Mutating K313 to alanine (black arrow marking K319A) abolishes the enzymatic activity of ALG11, so we expected E312G to have a strong growth defect.
Turning next to M408R, the previously published data on nearby amino acids was less clear as to what to expect. M408 (yellow arrow) is next to another evolutionarily conserved stretch of ALG11. However, mutating E413 (black arrow marking E413A) alone to alanine did not appear to affect ALG11 function. To be clear, M408 was not mutagenized in the original study so we have no real basis for making any predictions.
As we learned in the PGAP3 project, we can successfully pivot from patient-specific avatar to generic knockout avatar for the drug repurposing screens. That said, the endogenous ALG11 promoter plasmid is being constructed by Kelly and Tom so we can make one final attempt at revealing a growth defect in the ALG11 E312G and M408R yeast avatars.
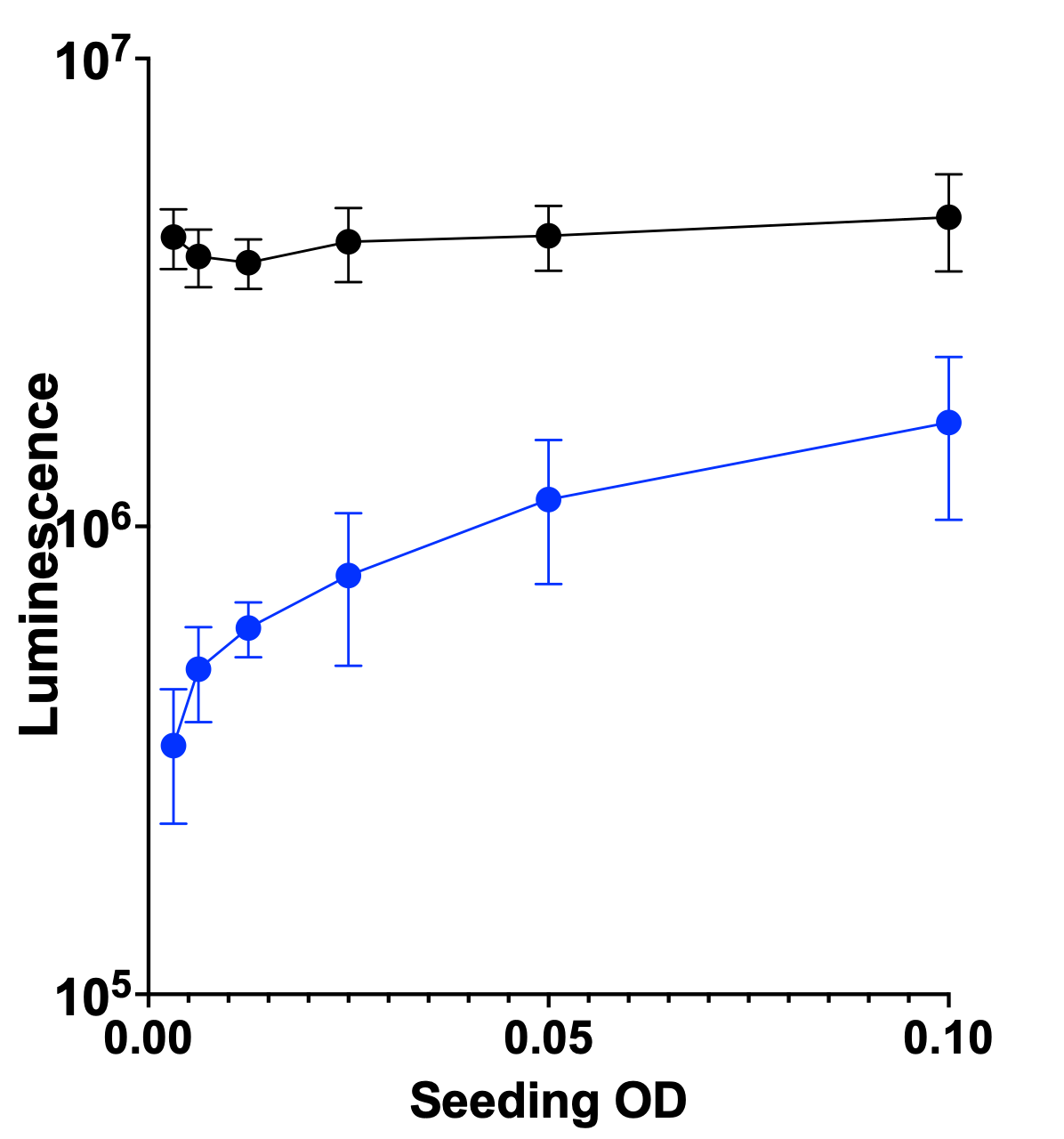
We also decided to advance the ALG11 KO yeast avatar based to the Pharmakon screen. Here’s the seeding density results at 24 hours at 37˚C. The wildtype is the black circles and line; the ALG11 KO yeast avatar is the blue circles and line.
Here’s the Z’ optimization experiment showing about as night-and-day a contrast between wildtype and mutant that we’ve seen.
Next step: Pharmakon screen. Stay tuned!