Decentralized yeast lab for drug repurposing
Perlara is renting a bench at SFBiolabs and staffing it with yeast experimentalist Cure Guides from UCSF. Our pop-up yeast lab went live last month, and the first project out of the gate is PGAP3-CDG.
Encouraged by the milestones of the ongoing UCSF yeast avatar drug repurposing project, we decided to pop up our own low-cost, low-complexity, yeast-powered drug repurposing lab outside of academia, thereby filling a gap in the research services market not currently filled by any contract research organizations (CROs). We recruited a junior specialist from the UCSF project as well as two grad students from yeast labs, assembling Perlara’s first distributed team of experimentalist Cure Guides working on behalf of a Bay Area family foundation called Moonshots for Unicorns.
Moonshots for Unicorns is the family foundation partner driving this research forward for Lucy and every person affected by the ultra-rare genetic metabolic disease called PGAP3-CDG, one of over 100 known Congenital Disorders of Glycosylation. You can read up more on PGAP3-CDG and other CDGs on the wonderful resource CDG Hub.
The goal of this project is to identify a PGAP3-CDG drug repurposing clinical candidate using a rapid turnaround, yeast-powered phenotypic screening approach. It starts with the development and phenotypic characterization of genetically personalized yeast avatars followed by growth-rescue drug repurposing screens, a process previously published for PMM2-CDG yeast avatars in Lao et al., 2019 that resulted in the discovery of epalrestat and formation of Maggie’s Pearl. A long-awaited 40-patient Phase 3 clinical trial of epalrestat for the treatment of PMM2-CDG will soon begin at the Mayo Clinic in Minnesota.
The yeast gene most closely related to human PGAP3 is called PER1. We knew from the Saccharomyces Genome Database that a per1∆ whole-gene deletion mutant is viable but slow-growing and temperature sensitive, meaning its slow growth is further slowed by incubation at the excessively balmy 37˚C versus the preferred 30˚C. Slow growth can be rescued by small molecules in unbiased high-throughput screens where thousands of FDA approved (or near-approved) drugs are tested in parallel. We were able to purchase off-the-shelf per1∆ knockout-strains from the commercial vendor Horizon Discovery to go from cold start to 96-well liquid growth assay data in two weeks flat from August 1-14.


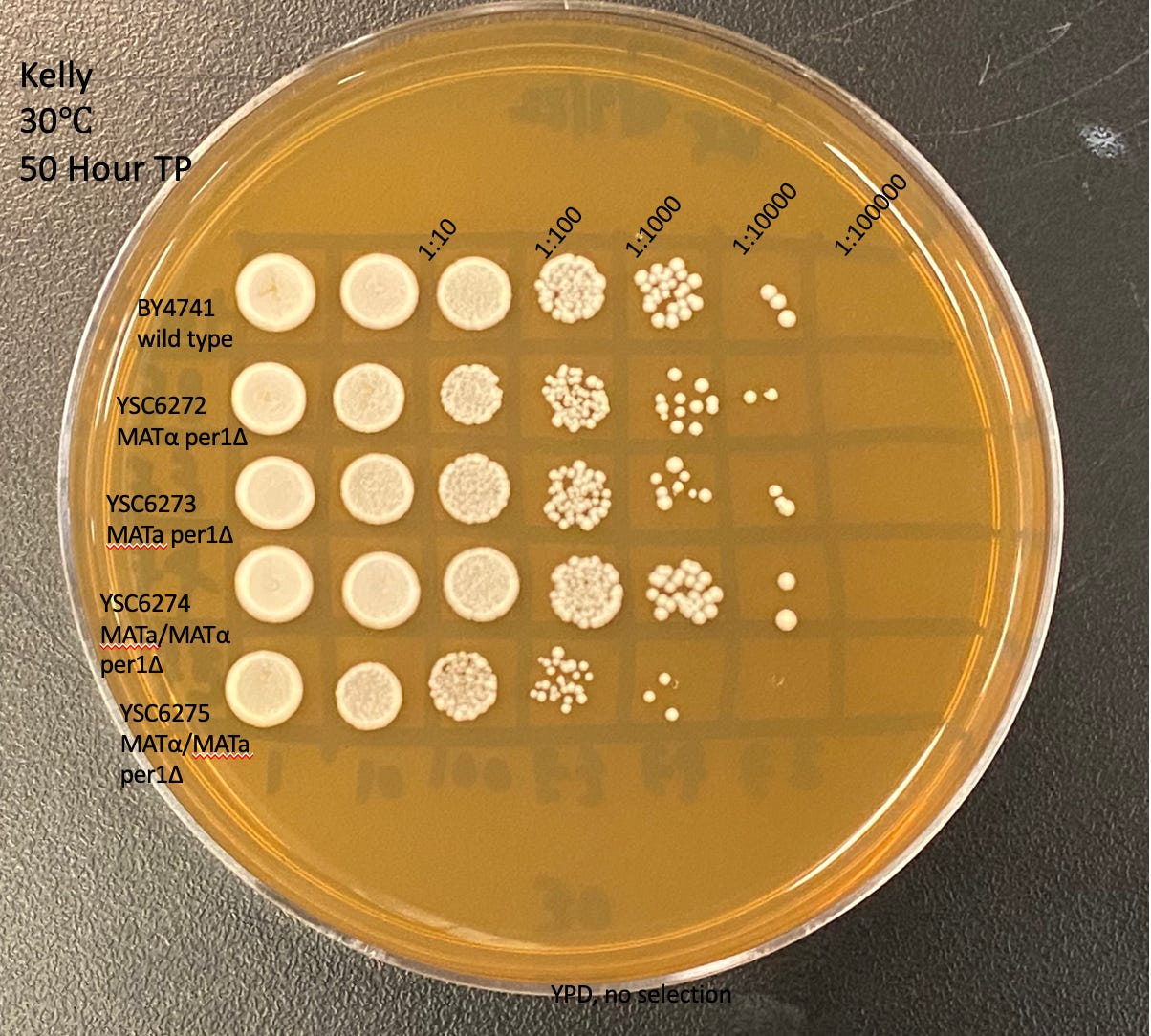
As we described previously, a yeast growth-based assay with appropriate positive and negative controls will be designed and optimized for 384-well plate drug repurposing screens. Yeast Cure Guides Kelly, Dana and Henry performed spot assays to assess growth differences on solid medium (agar plates) at either 30˚C or 37˚C.
Dana and Kelly performed independent technical replicates and got nearly identical results. At 30˚C, the two haploid yeast per1∆ mutants as well as the homozygous diploid per1∆/per1∆ mutant exhibit a modest (less than 10-fold) growth defect compared to wild-type isogenic control strains. As predicted from SGD, per1∆ mutants are sicker when grown at 37˚C. The homozygous diploid (YSC6275, bottom row) is several orders of magnitude more sensitive to high temperature compared to the heterozygous diploid control (4th row). Interestingly, the MATalpha mutant (YSC6272, 2nd row) appears to be more temperature-sensitive than the MATa mutant (YC6273), 3rd row).
In the second half of August, the Yeast Cure Guides switched from low-throughput assessment of growth on solid medium to medium-throughput assessment of growth in liquid medium in 96-well plates. The final assay optimization stage is happening now, where Z-scores will be assessed this week in high-throughput 384-well-plate format at UCSF Small Molecule Discovery Center, followed by a screen of the first drug repurposing library on deck. The results from the spot assay on petri dishes replicated robustly in liquid media, as shown in the summary growth curve plot below.
By mid-September, we expect to have data from the per1∆ mutant screens, just in time to receive plasmids from Twist Bioscience that will allow us to create a Lucy yeast avatar expressing her missense variant. The growth of the Lucy yeast avatar will be assessed at 37˚C and compared to the per1∆ whole-gene deletion strains. We expect Lucy’s mutation to be hypomorphic, meaning there is residual PGAP3 activity that could be potentiated or bypassed by the actions of a known drug.