Drug repurposing for PACS2 with Cell Painting
The CureMap we created for PACS2 Research Foundation recommended drug repurposing that combines patient fibroblasts and an unbiased image-based phenotypic screening approach called Cell Painting.
In collaboration with
We published a PACS2 CureMap for the PACS2 Research Foundation last winter. Our top recommendations included developing E209K disease models and high-throughput phenotypic assays for drug repurposing screens. The most actionable and cost effective disease model is a primary patient-derived fibroblast line established from a ~3mm skin punch biopsy that could be performed by a local dermatologist. We also encouraged iPSC line development, including an isogenic corrected control as part of an allelic series. Brain organoids, worm and fly avatars are on the menu, too.
In this update, the focus is on Lena’s fibroblasts and Cell Painting, a technique originally developed in 2016 by the Carpenter-Singh Lab at the Broad Institute of Harvard and MIT aka The Broad. As the initial essential quality control step by the team of scientists working on this project at an established CRO, the heterozygosity of the E209K pathogenic variant in Lena’s and her sister’s fibroblasts was confirmed by PCR below.
Previous drug repurposing screens of patient-derived fibroblasts involved a challenge assay. For example, growing mitochondrial disease patient fibroblasts in glucose and then abruptly switching to galactose, thereby forcing the cells to use their mitochondria to survive. If their mitochondria can’t function properly, the cells die. A challenge assay could also involve stressing cells with excess of a compound they can’t metabolize. That seemed like a reasonable direction to go at the outset given that PACS2 protein is enriched in so-called mitochondria-associated membranes (MAMs).
Yet given how much is still unknown about PACS2 function in a cell let alone a human, focusing on the mitochondria or the ability for a cell to respire introduces bias. That’s why we turned to Cell Painting. The premise of Cell Painting is that there is no one cellular phenotypic assay to rule them all when you don’t understand the function of the protein mutated in an affected person. The multi-organellar presence of PACS2 combined with keeping the mouth of the funnel as wide as possible made Cell Painting a logical fit.
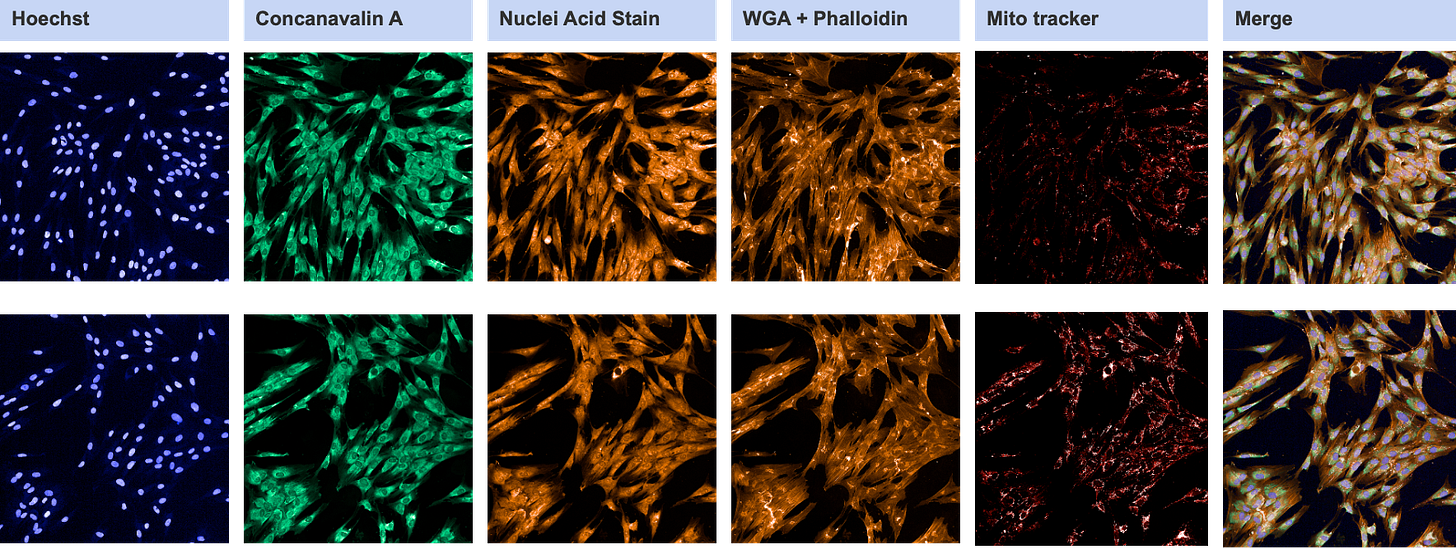
Cell Painting is a mixtape of vital dyes, stains that indicate the presence and boundaries of intracellular organelles:
Hoechst: nucleus
Concanavalin A: endoplasmic reticulum (ER)
RNA Stain: cytoplasm, nucleolus
WGA: Golgi
Phalloidin: actin cytoskeleton, plasma membrane
MitoTracker: mitochondria
Here’s the merged image. Can you tell by eye that they’re not only different but also all the tiny ways they’re different? The human eye is not great at this task but computer vision was made for it.
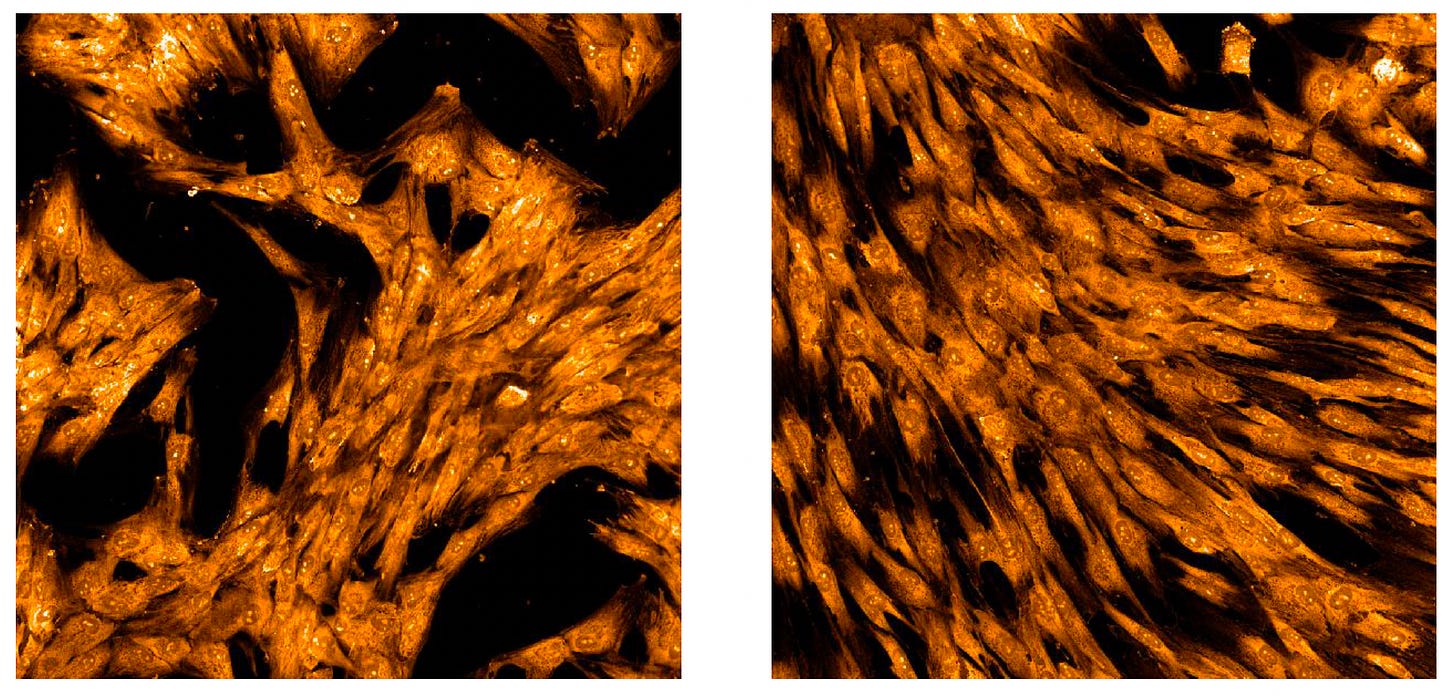
Let’s play the mixtape and listen carefully to each track. In the sequence of paired images below, the patient cells are on the left and the control cells are on the right. See if you can pick up on the differences, any differences.
First up: the cell nucleus stain Hoechst. If this had been a fibroblast from a patient with a pathogenic lamin variant, the nuclei on the left would be crescent-shaped or elongated depending on the exact mutation.
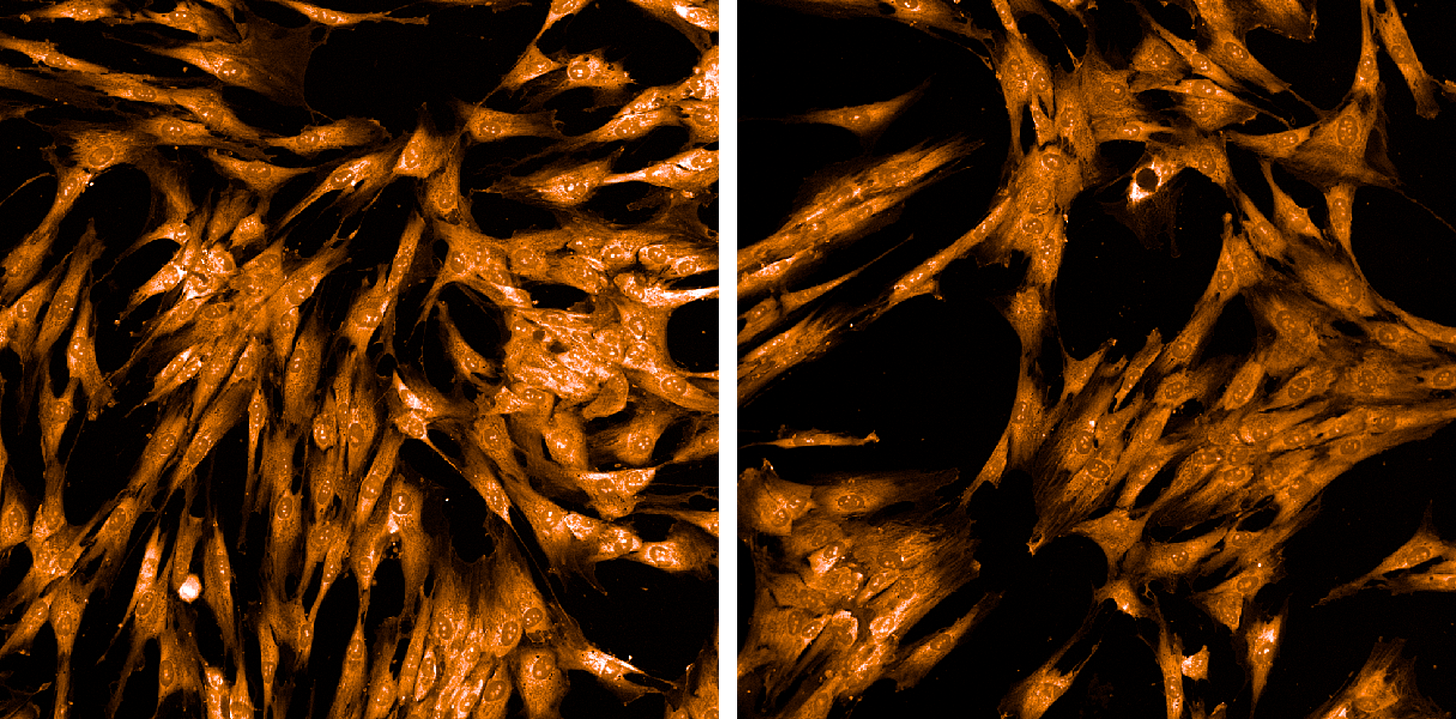
Next up: Concanavalin A/Alexa Fluor 488 conjugate, a fluorophore-tagged carbohydrate-binding protein that binds to the mannose residues coating proteins in the endoplasmic reticulum (ER).
Next up: SYT0 14 green fluorescent nucleic acid stain, which marks nucleoli and cytoplasmic RNA.
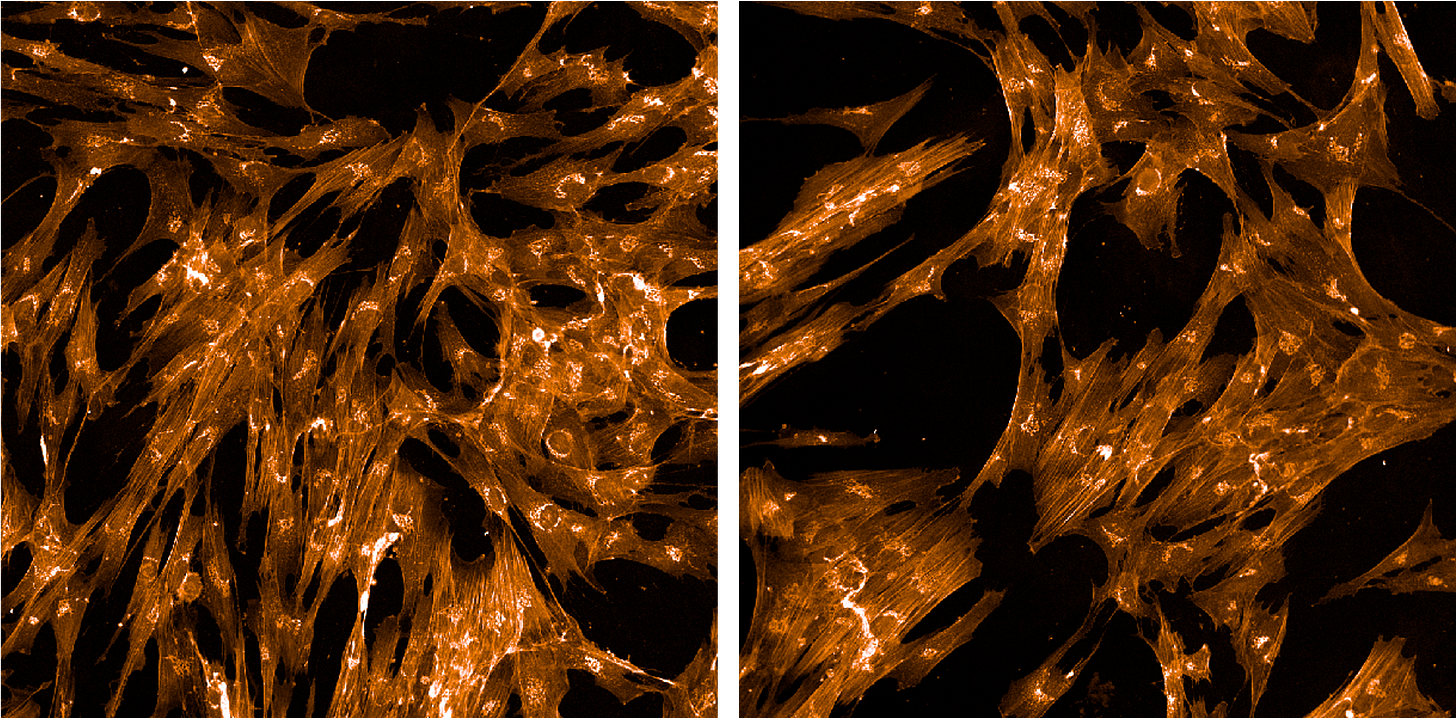
Next up: Phalloidin/Alexa Fluor 568 conjugate and wheat-germ agglutinin/Alexa Fluor 555 conjugate. The phalloidin binds to the actin cytoskeleton. The WGA lights up the Golgi and plasma membrane.
Finally, we have Mitotracker Deep Red, which as its name suggests illuminates the whereabouts of mitochondria.
The playlist view with the patient cells on the top row and the control cells on the bottom row:
Images of thousands of cells were processed by well-established methodological workflows:
Cell segmentation: identification of (sub)cellular boundaries by image analysis software, e.g., what is a cell, what is a nucleus, …)
Feature extraction: calculation of ~4000 different features per cell, e.g., shape, intensity, texture, co-localization, …)
Data pre-processing: pre-selection of morphologically relevant parameters (~4,000 parameters reduced to ~200)
Statistical significance: 20-45% of features had significant Z-scores depending in the cell density and incubation length
Even at ~200 features, some capture more of the variance than others. Using a dimensionality-reducing computational method called principal component analysis (PCA), Lena’s fibroblasts are the (green) dots on the left and her sister’s fibroblasts are the (red) dots on the right. The goal is now to find repurposable compounds/drugs that move Lena’s cells closer to her sister’s in PCA disease space.
A summary of the significant features shows that each of the Cell Painting stains yields statistically significant Z-scores, not just the mitochondrial, ER, and Golgi markers that were expected based on the published (and unpublished) literature on PACS2 or extrapolations from PACS1 biology.
We’re currently in the assay validation stage, which includes testing a few compounds that are known to affect PACS2 biology. Next, we’ll conduct the actual drug screens. In anticipation of a successful drug screening campaign, efforts are underway to prepare to validate fibroblast Cell Painting hits in one or more of the following PACS2 disease models: iPSC-differentiated neurons, brain organoid, worm avatar, fly avatar.