Drug repurposing with yeast avatars of SURF1 Leigh Syndrome
We started working with Cure Mito in August on a yeast drug repurposing project based on the template of our previous work with UCSF on MEPAN. Last week we completed the first screen and we have hits!
We’ve known for over two decades that the mitochondrial disease called Leigh Syndrome is caused by loss-of-function mutations in the gene SURF1 that can be disease modeled in yeast. In partnership with Cure Mito Foundation — shoutouts to Kasey Woleben and Lauren Ashwin for spearheading this project — we decided to finally do something about the decades-old observation that yeast cells completely lacking SURF1 are unable to grow in conditions that require their mitochondria to produce energy. We just completed a pilot screen of ~2,500 known drugs and identified a unique hit signature.
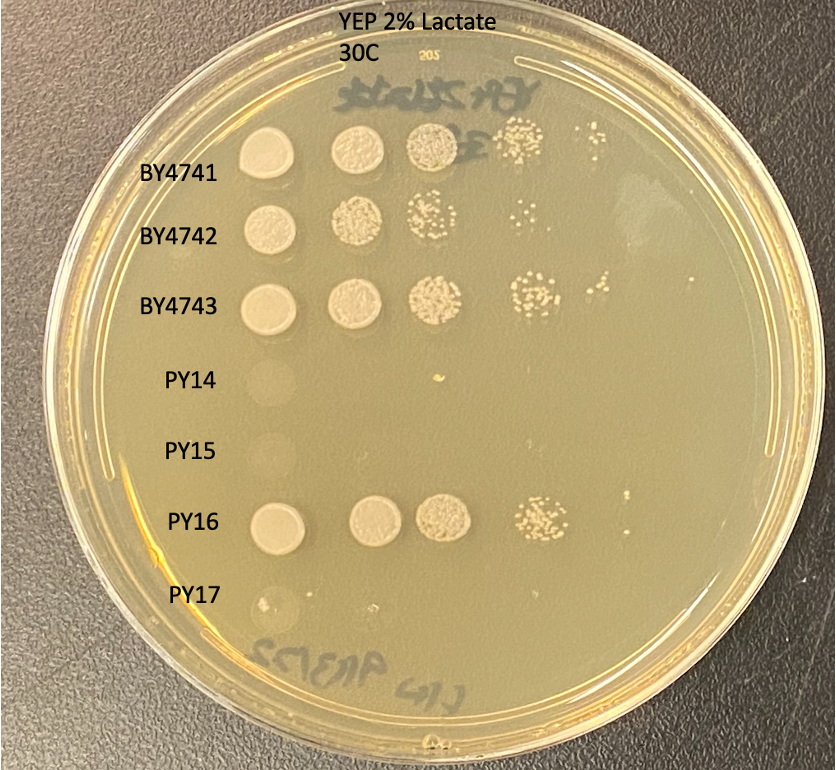
After completing the pilot Pharmakon PGAP3 yeast drug repurposing screen six weeks ago, the next program in the queue was for SURF1 Leigh Syndrome on behalf of Cure Mito Foundation. Fortunately (and unfortunately from a translational point of view) we had a massive head start based on results that were first published in 1997, when the first lab demonstrated that knocking out the SURF1 ortholog in yeast — SHY1 — results in loss of the ability of the knockout mutant to grow on a non-fermentable carbon source, e.g., lactate or glycerol, that requires functional mitochondria. A followup paper published by another group in 2002 used a spotting assay to demonstrate that a shy1∆ knockout mutant is incapable of growing on YEPG, or yeast media spiked with ethanol and glycerol as the sole non-fermentable carbon sources.

20 years later, we not only reproduced those results at our first yeast repurposing popup “cure lab” located at SFBiolabs, as shown below, but we also took the logical next step and performed the drug repurposing screen that could have been done years ago. BY4741 and BY4742 are wildtype haploid controls; BY4743 is the wildtype diploid control. PY14 and PY15 are haploid shy1∆ knockout mutants of each yeast mating type. PY16 is the diploid SHY1/shy1∆ heterozygous mutant, which grows just like BY4743. PY17 is the diploid homozygous shy1∆ knockout mutant.
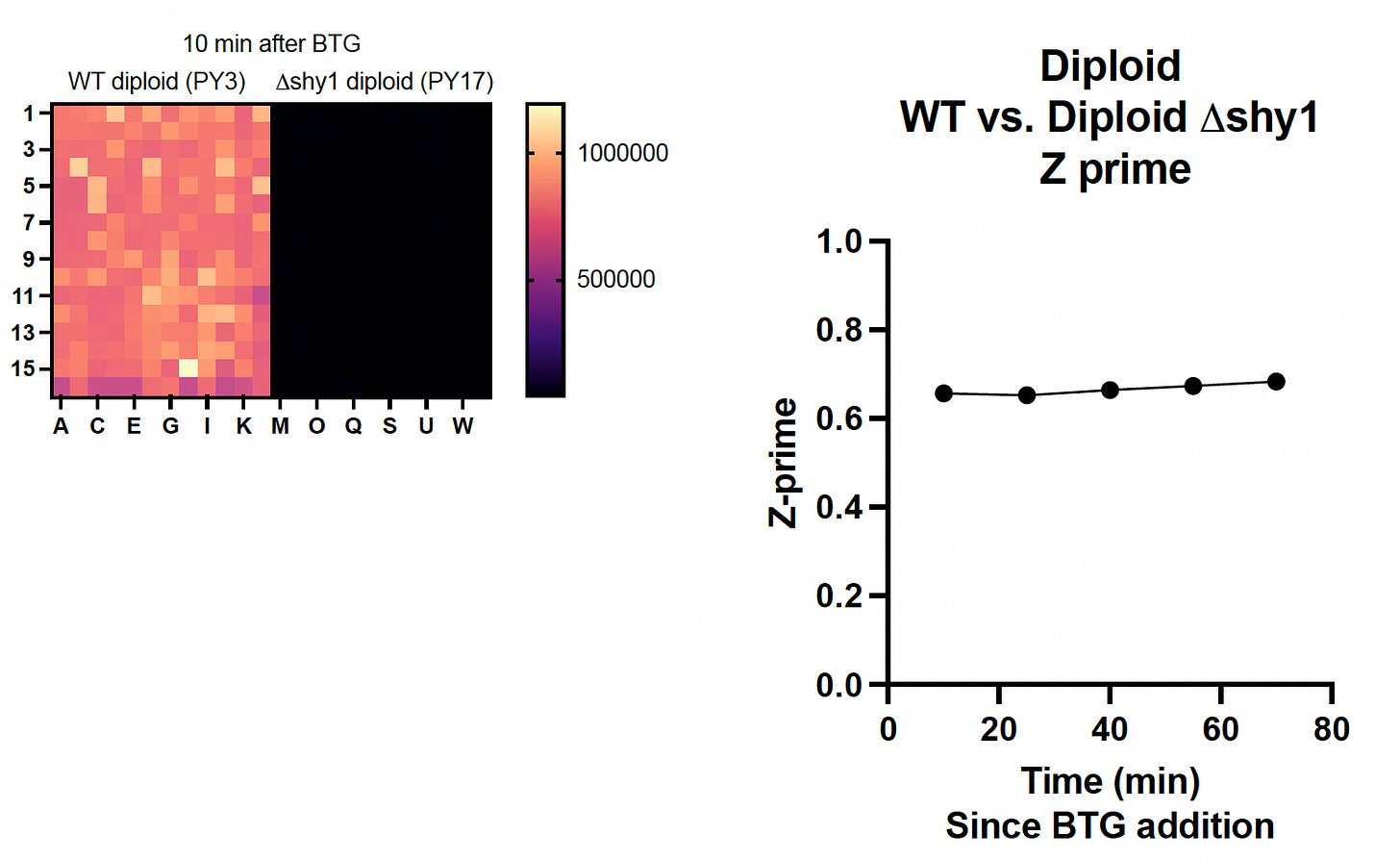
In stage two of our increasingly containerized (and therefore productized) yeast drug repurposing workflow, we transition from spotting assay on solid medium in petri dishes to a Bac-TiterGlo luminescence assay in liquid media in 96-well plates. The minimal growth phenotype we observed on solid media was perfectly replicated in liquid media, as shown below. Note that the diploid positive control BY4743 grows faster than the haploid positive control BY4741. Similarly, the diploid shy1∆ knockout mutant grows a tad faster than the haploid shy1∆ knockout mutant.
In stage three of our modular yeast drug repurposing workflow, we scale up from 96-well plates to 384-well plates for the pivotal Z’ optimization experiment, which is the last step before we advance to a pilot drug repurposing screen of the Pharmakon library. The typical Z’ cutoff is 0.5, which was breezily exceeded by both the haploid and diploid homozygous shy1∆ knockout mutants.
Because the Z’ was sufficiently high and because the large absolute difference in luminescence units (cell number) between the positive control and the shy1∆ knockout mutants at baseline, intensity b-scores (equivalent to a Z-score) soared into the 100s, yielding approximately 50 hits that resulted in at least a doubling of the number of shy1∆1 knockout mutant cells compared to untreated (placebo) mutant cells.
Here’s the full dataset showing the ~2,500 library compounds (red triangles) plotted along with the positive controls (cyan triangles) and the negative controls (green triangles) that are obscured by the baseline of inactive compounds at 0 intensity activation (cell number). The discernible red triangles are the hits; the larger the triangles, the more statistically significant the result. Remarkably, two compounds resulted in growth by shy1∆ knockout mutants that exceeded or nearly exceeded the growth of the positive controls.
As we pore over the data in the days and weeks ahead with our partners from Cure Mito, we’re excited to share more. In the meantime, I’ll offer this teaser. We see overlapping hits between these shy1∆ knockout mutant screens and the MEPAN screens from the UCSF collaboration, which we posted about here. More to come soon..








Can Bezafibrate be a repurpose drug for Surf1?