GNAO1 yeast avatars for drug multipurposing
A systems engineer and rare dad in Spain reached out to us with the opportunity to revive an old PerlQuest as a globally distributed drug multipurposing project. Last week we got the first hits.
We began working officially with Emiliano and a small network of G203R families that comprise GNAO1 Action last year, but he first made contact in the summer of 2021 inquiring about ways Perlara 2.0 could help his fellow G203R families. We assembled a distributed yeast team with members located in New Zealand, the US (California and Michigan) and Spain. After over half a year of optimization, we finally got an image-based assay and a GNAO1 yeast avatar to work at the same time last month. We identified two structurally similar hits that will be retested in the lab this week.
The proverb “If at first you don’t succeed, try, try again” must have been first uttered by a scientist.
In 2018, Perlara 1.0 started a collaboration with the Undiagnosed Diseases Network (UDN) to develop GNAO1 yeast and worm patient avatars expressing the de novo variant A221D as described on the Perlara 1.0 blog here and here.
That was one of the last screens performed at Perlara 1.0 in the fading light of 2018. Then the company went into hibernation mode for two years during the winter of 2019. Tantalizingly, two well-known statin drugs were identified as presumptive screening positives but hit potency determination in yeast was not performed nor were hits validated as planned in other GNAO1 yeast avatars or more complex GNAO1 disease models, such as worm and fly A221D avatars.
The project went cold for a little over two years until the summer of 2021, when an Argentine from Spain named Emiliano Pontoriero filled out what was then a simple form for families interested in working with Perlara 2.0. Emiliano’s daughter Alba has the devastating G203R variant in the evolutionarily conserved GNAO1 gene. But she is not alone in her fight!
G203R mouse models exist but disease phenotypes are mostly behavioral, expensive to assess, and require the expertise of specific labs, which practically means only a few repurposing candidates can be tested. The neuron-specific functions of GNAO1 and the varying effects of GNAO1 in GPCR signaling across the brain mean the translational potential of patient-derived fibroblasts is limited.
A G203R knock-in worm model (Wang et al., 2021) and a G203R/+ heterozygous fly model (Larasati et al., 2021) are described in the literature. The wall climbing defect of the G203R/+ fly model is well suited for low-throughput hit validation but not to a high-throughput screen in a traditional academic fly lab setting.
However, the G203R worm model described in Wang et al is a homozygous mutant; a G203R/+ heterozygous mutant was not mentioned in the paper, but a conversation with the publishing lab head (Prof Brock Gill) revealed that the G203R/+ heterozygous mutant was in fact generated but not explicitly described in Wang et al., 2021. Subsequent conversations revealed that the published G203R/+ heterozygous mutant was spurious and an attempt to replicate it from scratch has thus far failed, indicating that a G203R/+ heterozygous worm avatar may be a dead-end for now.
That left an opening for creating G203R yeast avatars.
Since last summer we’ve been working with Dr Andrew Munkacsi’s lab and recently minted PhD Dr Jeff Sheridan, both accomplished yeast chemical biologists at Victoria University in Wellington New Zealand. Yeast Cure Guide Dr Mathura Thevandavakkam is lead the project planning and project management.
As shown in the protein sequence alignment below, the ancestral version of GNAO1 in yeast is called GPA1. Glycine 203 in human GNAO1 corresponds to glycine 321 in yeast GPA1. In fact, the amino acid glycine is evolutionarily conserved at this position in all animals, not surprising as this region of the GNAO1 protein is involved in nucleotide binding:
We’ll spare the details but we tried several different knock-in and plasmid-expression approaches to reveal a screenable growth defect of G203R yeast mutant strains — frustratingly, all attempts ended in vain.
Along the way we encountered technical hurdles (expression plasmids that didn’t work as expected) and logistical delays (clearing customs at the New Zealand border is non-trivial).
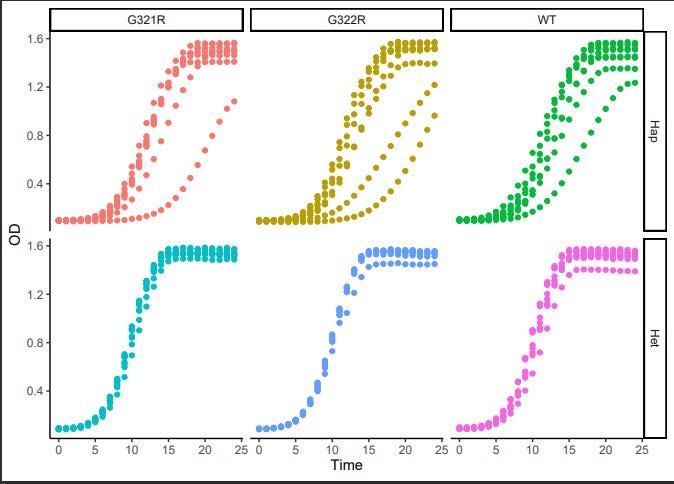
Below is a summary of growth curves of G321R and G322R yeast avatars where we engineered the mutations into the GPA1 locus in a haploid yeast genome. Expressing either G321R or G322R alone is not toxic.
Jeff also compared what happens when G321R and G322R are expressed in the heterozygous condition, mimicking the autosomal dominant genetic architecture seen in humans. As we observed with the haploid avatars, the heterozygous avatar has no observable growth defect.
Here’s Jeff walking us through the final growth-based screen we attempted just so you get a sense of the experimental workflow.
In consultation with yeast GPA1 expert Professor Henrik Dohlman’s lab, we pivoted from a generic growth-based assay to a pheromone-pathway-specific, image-based assay that directly reads out GNAO1/GPA1 function. Unlike humans with north of 800 GPCRs, the yeast GPCR repertoire is limited to receptors: a pheromone-sensing receptor and glucose-sensing receptor.
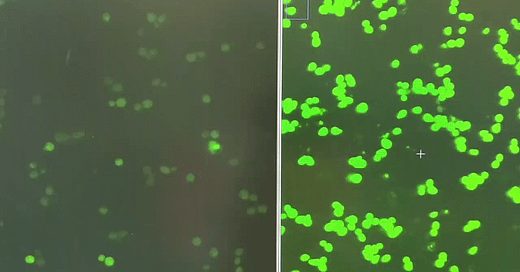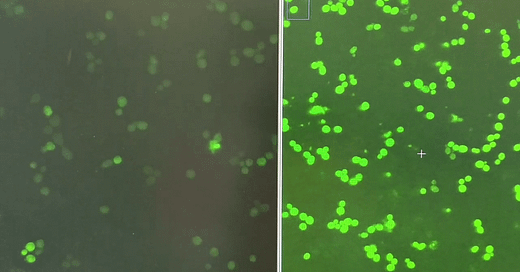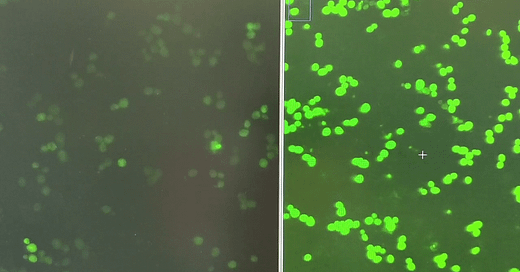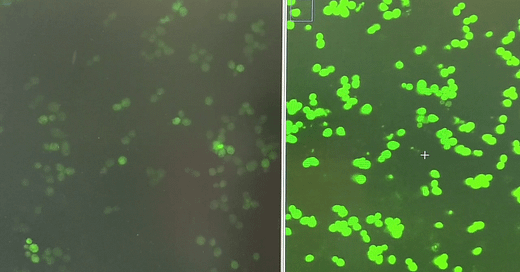
When yeast cells express the FUS1-GFP reporter, which is activated when yeast cells get a whiff of the mating pheromone called alpha factor, they light up like fireflies in midsummer.
Using plasmids battle-tested by the Dohlman lab, we measured the performance of yeast expressing G322A, which corresponds to G204, the glycine next door to G203, as well as other previously characterized GNAO1/GPA1 mutants in the FUS1-GFP reporter assay.
There was enough of a delta between G322A and WT (wildtype) that we felt confident to advance to screening the LOPAC and Spectrum Collection libraries.
Jeff’s first screen identified two structurally related hits that increase GFP expression in the first replicate of the screen. A second replicate will be completed this week. Flow cytometry will be used to validate hits at multiple doses on a single-cell basis, and also avoids the complication of cell clumping of pheromone-treated cells.
Of course the screen also identified enhancers, which will be informative in our interpretation of the suppressors, like the hit shown on the right compared to a control well on the left.
We’ll be able to speak more conclusively soon.