Landry's PMM2-CDG yeast avatar
There are 130 variants of PMM2-CDG that we know about, and the list is growing. We created a Landry yeast avatar expressing his combination of PMM2 variants and it's ready for 1-to-n drug repurposing.
In collaboration with
In 2017, we modeled three PMM2-CDG variants as part of Perlara 1.0's PMM2-CDG PerlQuest. We initially focused on recurring variants that were previously studied in the lab but never before in yeast: R141H, F119L and V231M. In 2018, we advanced F119L and V231M avatars to a proof-of-concept PMM2-CDG drug repurposing screen of 2,500 compounds.
Today we know of at least 130 PMM2-CDG variants. More PMM2-CDG variants, including so-called variants of unknown significance, will be discovered. It’s just a matter of time.
In this second PMM2 drug repurposing project, we created a new PMM2-CDG patient avatar with the goal of advancing it to a ~8,000-compound screen. This project will show us if there are known drugs beyond acetazolamide and epalrestat that could be repurposed for PMM2-CDG. We already published preliminary preclinical data from the original PerlQuest PMM2-CDG yeast avatars — the same ones that led us to the aldose reductase inhibition mechanism — that another CDG gene called PGM1 may be a disease-modifying drug target.
Like most kiddos living with PMM2-CDG, Landry is what geneticists call a compound heterozygote, meaning he inherited one pathogenic PMM2 variant from mom and different pathogenic PMM2 variant from dad. The yeast version of PMM2 is called SEC53. As shown in the protein sequence alignment below, the amino acid order is offset by +7 in yeast, so position 157 in human PMM2 is position 164 in yeast SEC53; position 101 in human PMM2 is position 108 in yeast SEC53.
The F157S mutation replaces an evolutionarily conserved phenylalanine with a serine residue and was predicted to be pathogenic in the genetic testing report. The N101K mutation was also predicted to be pathogenic but it is not evolutionarily conserved between humans and yeast. However, replacing either human asparagine 101 or yeast valine 108 with a lysine residue is predicted to be deleterious.
A recent paper characterized 15 known PMM2-CDG variants. N101 and F157 are both clearly in the cap domain, which is required for dimer formation and proper active site stabilization.

A flattened one-dimensional sequence of amino acid letters doesn’t do PMM2 justice. So we performed our own AlphaFold explorations. Here’s the predicted 3-D structure of the 246-amino-acid human PMM2 protein. Landry’s two variants are marked by the yellow arrows.
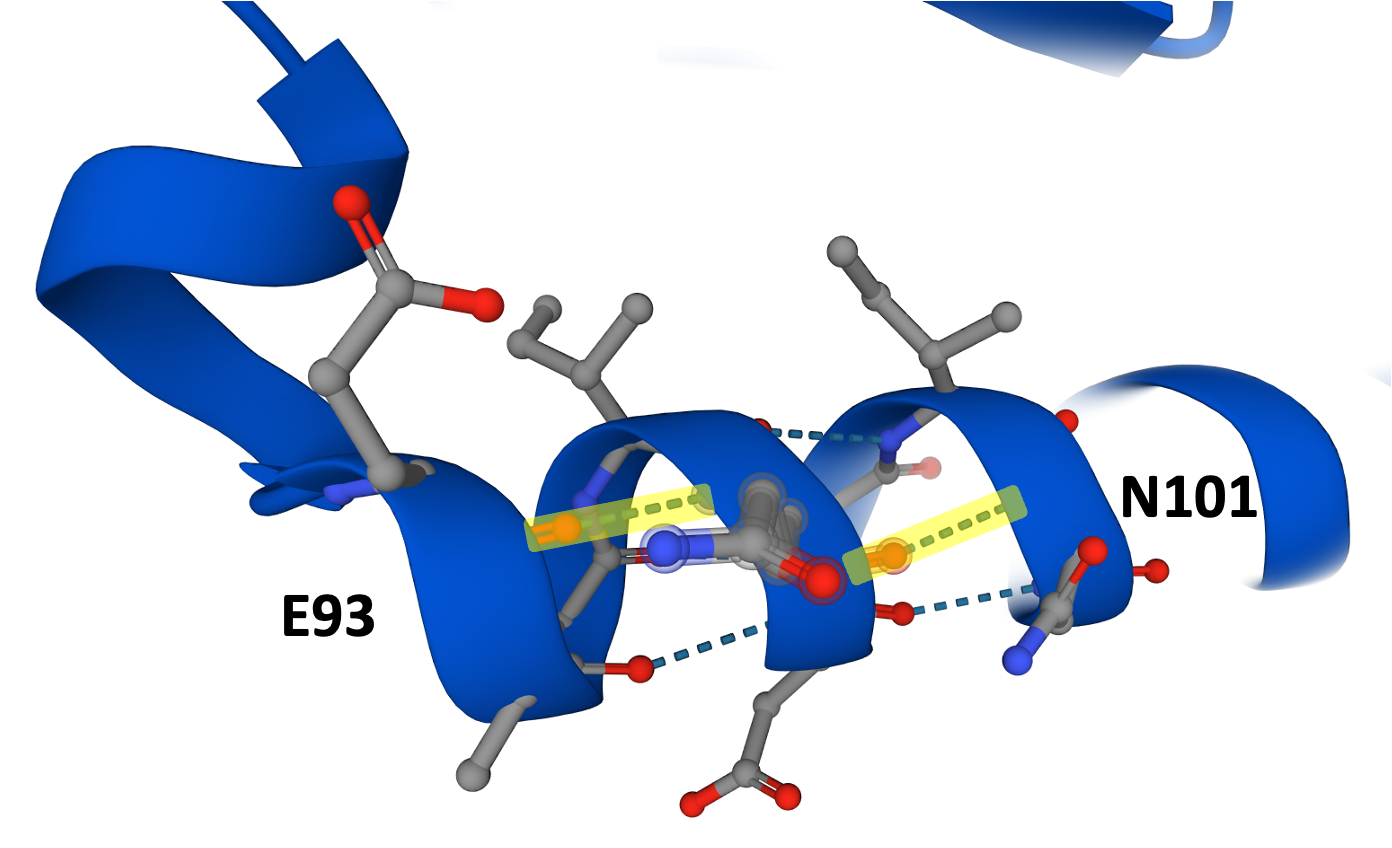
Let’s zoom in on N101 first. In the first PMM2-CDG yeast drug repurposing project where we modeled Maggie’s variants (E139K & R141H), we also created an E93 (glutamate 93) yeast avatar with no specific PMM2-CDG patient in mind. In our 2019 paper, we showed that the E93K avatar has a severe growth defect.
E93 and N101 are located in the same alpha helix that forms the dimerization interface between two PMM2 monomers. E93 participates in a hydrogen bond network with Q97 (glutamine 97), which in turn is hydrogen bonded to N101. So, we expected N101 to behave like E93: a severe loss-of-function.
Next let’s zoom in on F157. From the alpha helix below, phenylalanine 157 is high-fiving tyrosine 106, which is upside-down in this rendering. Tyrosine 106 is hydrogen bonded to tyrosine 102 within their shared alpha helix, and tyrosine 102 is next-door neighbors with N101, Landry’s other variant. That means in 3-dimensional space, both of Landry’s variants are indirectly interacting with each other.
AlphaFold is an amazing online tool but its prediction exist only in silico. Yeast avatars engineered to express Landry’s mutations are the fastest turnaround and most cost-effective disease models in which to characterize the N101K and F157S variants in the real world.
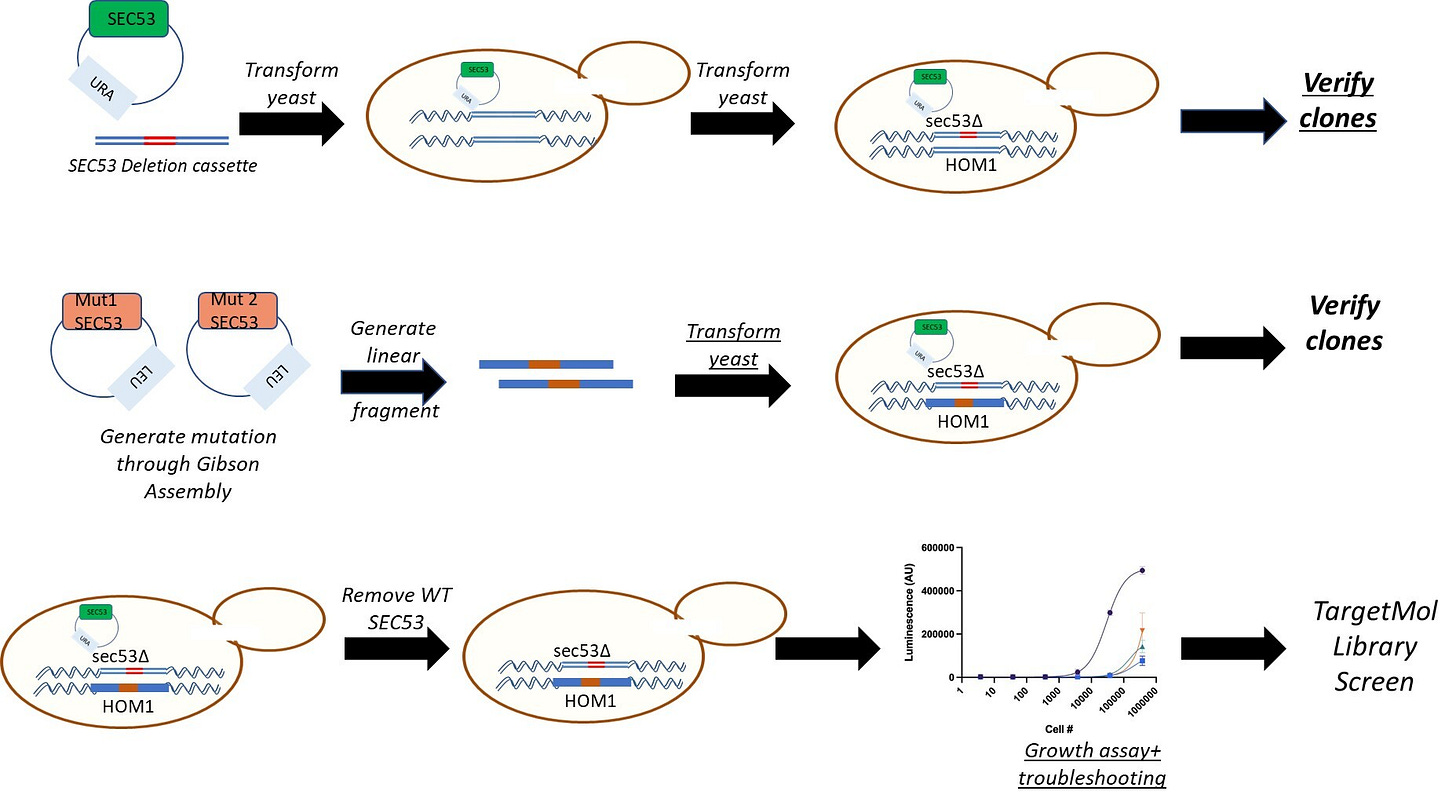
Here’s the one-at-a-time experimental workflow for generating a patient avatar that we first developed in the PerlQuest era. We didn’t want to introduce too many new variables so we could make an apples-to-apples comparison between this project and the first iteration.
Landry’s PMM2 variants were initially assessed as V108K and F164S haploid yeast avatars so we could assess their residual function separately. Then we would combine the two haploid avatars to create a diploid compound heterozygote, or compound het, that matches Landry’s exact genotype.
Both variants were computationally predicted to be damaging but the exact defect or
defects caused by each mutation – protein misfolding, protein instability, protein mis-
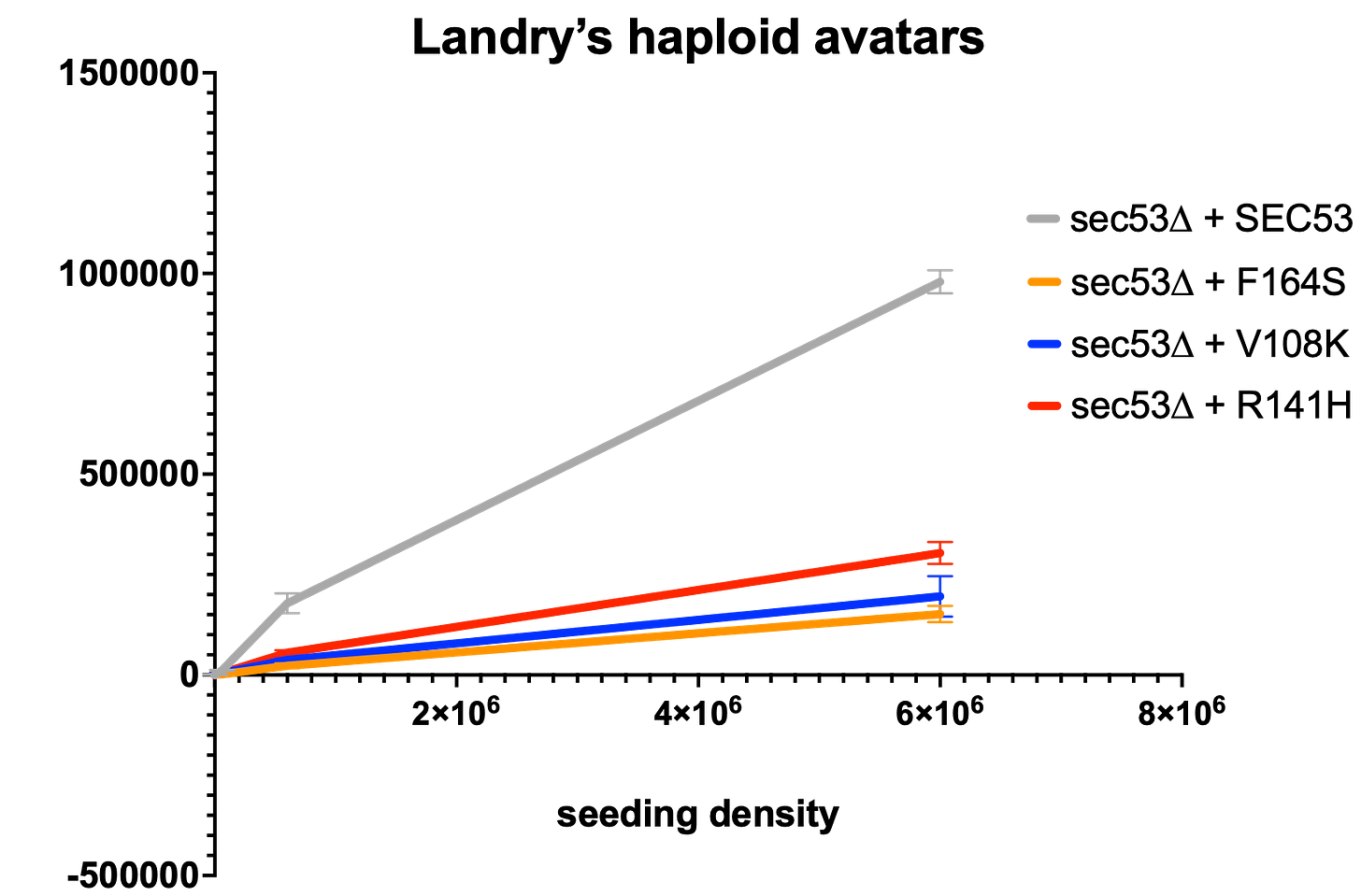
localization, catalytic site inactivation, among others – has not yet been elucidated. As shown below, each haploid Landry variant avatar has a growth defect as severe as R141H, which is considered a null, genetically speaking, meaning no detectable enzymatic activity.
The haploid avatar results suggested to us that Landry’s diploid compound heterozygous avatar would also have a severe growth defect. And that is indeed the case, as shown below.
We’re excited to advance the Landry’s compound het avatar to a ~8,000-molecule drug repurposing screen next month in collaboration with UC Berkeley and UC Santa Cruz.
Is there a more efficient and scalable way to generate PMM2-CDG yeast avatars that’s also more democratized than making and testing one or two variants at a time? We believe there is a way to create all possible PMM2-CDG yeast avatars in a one-and-done experiment. Stay tuned!