Drug multipurposing for PIGS-CDG using yeast
We began working with CDG CARE and several pioneer CDG communities last summer. PIGS-CDG is one of 170+ CDGs, most of which are ultra-rare. CDG stands for Congenital Disorder of Glycosylation.

The extraordinary degree of evolutionary conservation of glycosylation pathways across the animal kingdom combined with the extraordinary unmet medical need of CDG communities led last summer to the launch of several yeast-powered drug multipurposing projects, including one for PIGS-CDG. It took longer than expected to complete the assay development stage but we’re finally ready for the drug screening stage. Here’s a summary of the journey thus far.
Collaborators
Following discussions at the 2022 Rare Disease Day symposium & CDG/NGLY1 Family conference in San Diego, Perlara and several CDG pioneer communities decided to launch drug multipurposing projects modeled after the 2017-2018 PMM2-CDG PerlQuest that led to the discovery of epalrestat, which is currently in an actively recruiting Phase 3 study at Mayo Clinic.
A PIGS-CDG family with an affected daughter named Anna was one of those CDG pioneers we spoke to during the meeting. The PIGS gene encodes a protein that forms a part of the multi-subunit GPI transamidase complex involving the glycosylphosphatidylinositol pathway. Perlara Cure Guide and CDG Program Director Dr Kristin Kantautas nicely summarized what is known about the PIGS gene on CDG Hub here.
Patient Anna’s PIGS gene is compound heterozygous for a nonsense mutation and a duplication event. The nonsense mutation, W245X, replaces an evolutionarily conserved tryptophan residue with a termination codon likely resulting in a truncated protein.
The other mutation, an in-frame duplication p.Asp381_Val388dup in the PIGS gene, involves a methionine and arginine residue conserved between humans and yeast, whose PIGS ortholog is called GPI17.
These mutations are computationally predicted to be damaging but the exact defect or defects caused by each mutation – protein misfolding, protein instability, protein mis-localization, catalytic site inactivation, among others – have not yet been elucidated.
Yeast engineered to express Anna’s mutations are the fastest turnaround and most cost-effective disease models in which to characterize Anna’s W245X and p.Asp381_Val388dup variants, so we set out to generate Anna avatars and assess their growth.
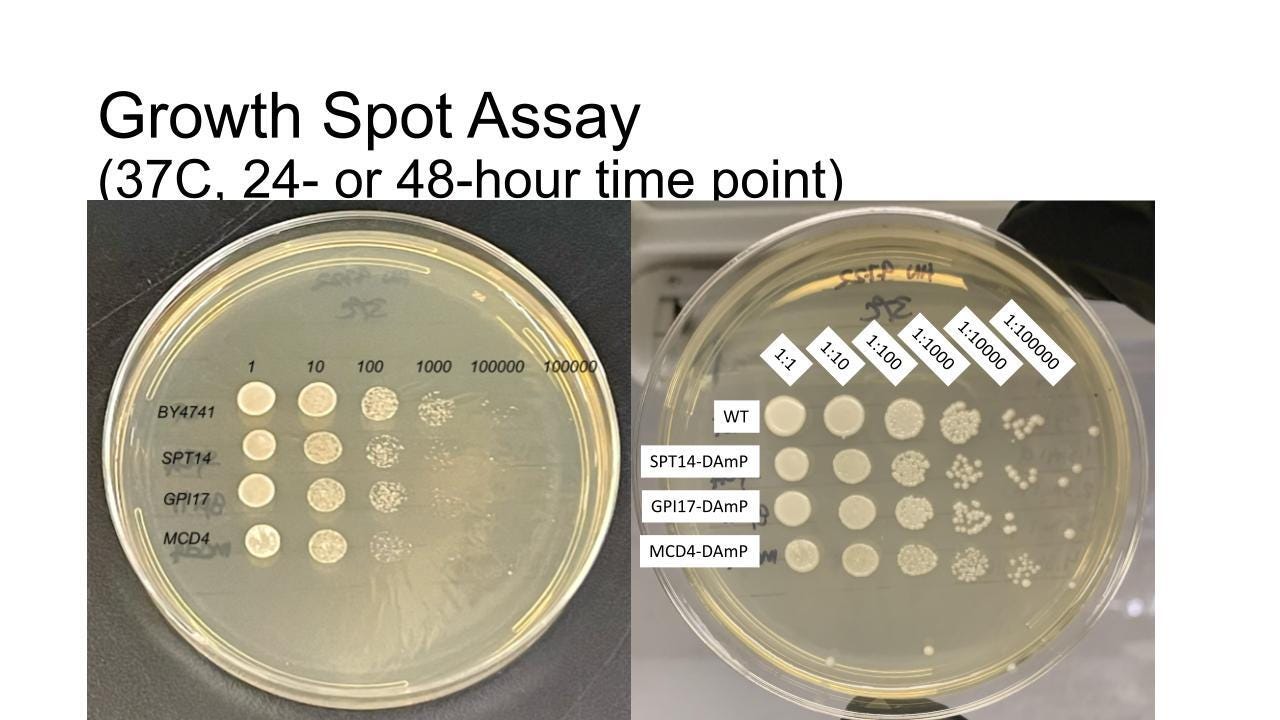
As a backup strategy, we ordered the GPI17 DAmP haploid strain which contains ~10% of normal GPI17 expression and would therefore serve as a generic loss-of-function mutant that in theory would behave like Anna’s avatars in the event that said avatars were not amenable to drug screening.
It proved to be a wise decision as we encountered challenges with avatar construction and other unexpected technical hurdles like flocculation that turned what we thought would take several weeks into several months of troubleshooting and protocol pivots.
Initial experiments (upper image) aimed at showing a growth defect at 30˚C did not work. Unlike our experience with PGAP3 where we exploited a growth defect at 37˚C, we weren’t able to observe a robust enough difference in growth between the GPI17 DAmP mutant as compared to wildtype.
We realized we needed to add a sensitizing agent to widen the delta between mutant and wildtype. After experimenting with high temperature and agents that cause cell wall stress, e.g., calcofluor white, we settled on hygromycin, an aminoglycoside antibiotic.
We’re planning to complete the final Z’ optimization experiments in the next 7-10 days, followed by the Pharmakon screen at SMDC in mid-February. Stay tuned for the next update in a few weeks.