MAN1B1-CDG Cure Odyssey
Two years ago a family launched a drug repurposing project. After failing with fibroblasts and worms, we got hits using a fly model last summer, which led a 1-to-N pioneer study currently in progress.
In collaboration with
We were introduced to Drs Claire Fast, MD and Matt Carroll, MD, parents of Jake and his brother Thomas, in early 2021. Claire and Matt knew how patient avatars led to discovery of epalrestat for PMM2-CDG. They hoped that patient avatars for MAN1B1-CDG might lead to targeted repurposing candidates.
The MAN1B1 gene encodes an ancient enzyme called mannosyl-oligosaccharide 1,2-alpha-mannosidase. Below is an illustration from the MAN1B1-CDG page on CDG Hub, an invaluable starter kit for any CDG family seeking aggregated foundational knowledge about their CDG gene.
Analogous to NGLY1 deficiency, where a glycanase that snips sugar chains from proteins is missing, MAN1B1-CDG is caused by deficiency of a mannosidase that trims a terminal mannose from one of the glycan’s antennae (shown below in the center of the diagram). Also similar to NGLY1, the prevailing wisdom is that MAN1B1 is part of the endoplasmic reticulum associated degradation or ERAD pathway, a one-way ticket to the cell’s protein recycling plant aka proteasome.
In other words, MAN1B1 is thought to play a generalist role, with no particular preference or responsibility for specific glycoproteins. It’s curious that the human genomes encodes so many closely related alpha-mannosidases like MAN1A1, MAN1C1, MAN1A2, MAN2B1, etc.
Could MAN1B1 actually be a specialist or have functions in addition to its enzymatic activity? According to AlphaFold, the C-terminus — where Jake’s mutations are located — comprises the mannosidase catalytic site. However, the N-terminus of MAN1B1 is terra incognita: a transmembrane domain is predicted, as well as multiple unstructured segments. Both of Jake’s variants are buried in the compact ball that is the C-terminus.
Only one way to find out: make patient avatars/disease models, stress test them, and then screen at least one model to identify drug repurposing hits. Keep in mind that the state of MAN1B1-CDG disease modeling in early 2021 resembled the state of PMM2-CDG disease modeling in early 2017. Besides patient fibroblasts and failed attempts at viable mouse models, no one was really exploiting invertebrate models except for Prof Kendall Broadie who developed a PMM2 fly model. "Perlara 1.0” created the first PMM2-CDG yeast avatars and PMM2-CDG worm avatars, and we were off to the repurposing races.
Claire and Matt launched JDC Research Co and the MAN1B1-CDG cure odyssey got underway.
Jake is compound heterozygous for two missense mutations at positions located in the enzyme’s catalytic region that are evolutionarily conserved in all animals, including yeast: R597W and F659C. These mutations are computationally predicted to be damaging but the exact defect or defects caused by each mutation – protein misfolding, protein instability, protein mis-localization, catalytic site inactivation, among others – has not yet been elucidated.
For MAN1B1-CDG, like for PMM2-CDG, we had the full manifest of Noah’s Ark of avatars to choose from: yeast, worm, fly, frog and zebrafish, oh my!
We initially focused on yeast since it’s usually the simplest and cheapest option. The yeast ortholog of MAN1B1 is MNS1. The immediate challenge was that a mns1∆ whole-gene deletion mutant is viable. That meant we’d have to figure out a way to sensitize the mns1∆ mutant with a mutation in a second gene or add an exogenous chemical stressor to reveal a screenable phenotype.
We were considering yeast a year before we popped up our first yeast cure lab, and so the cost of yeast model creation and characterization in an academic lab did not fit within the budget. We also had to maintain optionality for worms, flies and fibroblasts since we didn’t know a priori which disease model would make it to the drug screening stage.
Some genes don’t make our jobs easy. Worms encode four different orthologs of MAN1B1. We knew that they weren’t completely interchangeable in the worm because each MAN1B1 ortholog has a different spatial and temporal expression pattern. So we worked with In Vivo Biosystems to generate a series of knockouts, starting with a single knockout of mans-4 which eked out mans-3 as the closest MAN1B1 ortholog:
Single: mans-4 knockout (KO)
Double: mans-4/mans-1 KO
Triple: mans-4/mans-2/mans-1 KO
Quadruple: mans-4/mans-3/mans-2/mans-1 KO
If we didn’t see growth or behavioral phenotypes (deficits) in the mans-4 KO, we’d escalate to the double KO, escalate further to the triple KO, and only resort to the quadruple KO if none of the other knockouts yielded a phenotype. Unfortunately, none of the homozygous KOs exhibited inviability or sterility of reduced brood sizes.
This result left us in a situation not too dissimilar than what we found ourselves with the PMM2-CDG worm model back in 2018 at Perlara 1.0. It also didn’t have an overt phenotype when unstressed. We reasoned that the worm MAN1B1 KO’s would be more sensitive to tunicamycin or bortezomib. Tunicamycin is a N-linked protein glycosylation inhibitor. Bortezomib is a proteasome inhibitor that we previously used to sensitize NGLY1 and PMM2 worm avatars.
But we had to triage the worm program to stay under budget and reserve funds for a fly screen. We took a diversified approach to disease modeling because we knew that it’s possible to reach a dead-end when a model fails to yield screenable phenotypes.
Anticipating that we’d need to pivot to a cell-based model as a hedge in case all invertebrate models failed, we launched a fibroblast project with Charles River (CRL) at the same time as the worm project. What’s more, within the fibroblast project we put in place two complementary strategies: an enzyme activity assay and an image-based assay.
Again, we were influenced by our experience with PMM2-CDG, where we used a low-throughput biochemical PMM2-CDG enzymatic activity assay to validate hits from yeast and worm screens. We reasoned that we could use existing commercially available alpha-mannosidase kits and scale them up to 384-well format. In November 2021, we hit a wall here when it became clear that two off-the-shelf alpha-mannosidase assays weren’t intrinsically sensitive enough to get a meaningful signal. They required too many cells per well than was feasible.
Based on the Rymen et al paper from 2013, we pivoted to an image-based screen using Jake’s fibroblasts where we rescued a Golgi fragmentation phenotype. We asked CRL to test a panel of Golgi markers/stains on Jake’s cells as an age-matched control line in parallel.
By the end of December 2021, we had these tantalizingly encouraging preliminary results showing that two Golgi markers — TGN64 and WGA — distinguished between wildtype and Jake’s fibroblasts.
Let’s zoom in closer on TGN64 and WGA. The TGN64 signal appears to be more concentrated around the nucleus, leading to a lower density and total area of signal across each cell.
You can observe the same pattern with WGA.
Buoyed by what finally seemed like screenable results, we wanted to be completely certain of the results. We set out to replicate the TGN64 and WGA results in Dr Eva Morava-Kozicz’s lab at Mayo Clinic. A visiting medical student named Diederik DeGraef performed multiple careful replication studies in May and June 2022.
Unfortunately, we weren’t able to replicate CRL’s findings. We knew time of incubation mattered. We saw the December 2021 results after a 24-hour incubation but the effect was gone at 48 hours. We tried different fixation techniques but none of them seemed to make a difference.
Frustrated but not deterred from our ultimate goal of identifying a repurposable drug for Jake and other kiddos living with MAN1B1-CDG, we had one more ace up our sleeve: a fly model.
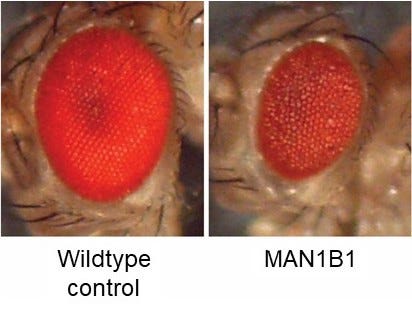
In March 2022, we launched a drug repurposing screen in Prof Clement Chow’s lab at the University of Utah. Knocking down MAN1B1 expression just in the fly eye results in a “rough eye” phenotype. The goal of the screen was to find drugs that make the unhealthy eyes on the right look like the shiny robust eyes on left. The fly eye screen is a cell-based phenotype (growth and survival) in the context of a living animal that has to consume the drug in its food.
Last summer, the Chow lab powered through the Prestwick Collection of ~1,500 compounds. By early November 2022, hit validation studies on the top class of hits were finished. 30% of hits belong to a single pharmacological class!
Six members of the class are shown below. Statistical significance is marked by the red asterisks. There appears to be dose-limiting toxicity that cancels out the efficacy at the highest dose. But there are also two examples (upper and lower left graphs) the display a dose response and have maximum efficacy at the highest dose 25µM.
Like all worthwhile science projects, we had to persevere through negative results, and not get disillusioned by false positive results or results that don’t replicate in independent labs. There were moments during the past two years when it felt like we’d exhausted all options, at least those with the operating budget. So when we had data that pointed to a well-known drug class including members that have been safely given to children, we stood at a crossroads that all rare disease parents and communities dream of: a treatment option.
On November 15, Jake became a CDG pioneer, joining Maggie from PMM2-CDG, Lucy from PGAP3-CDG, and there will be many more who will join them soon.

Here’s how Jake’s mom Claire summarized Jake’s first three weeks on drug:
The differences:
- he has been waking up earlier every day since the day he started the drug (not good!) - usually waking at 5:15 when he used to sleep to 5:45 or 6 - despite this he doesn't seem more tired during the day
- he is more playful and seems to engage with people more. He more often wants to tickle, wrestle, dance with us. He is clearly happy to see me when I come home which was inconsistent before despite being anxious when I leave.
- his aide, who doesn't know what we're doing, told me yesterday that he has been less aggressive during her lessons with him since we came home from vacation (when we started the drug). In the fall, unfortunately, he had been pushing and sometimes hitting when he was frustrated or didn't want to do something. She says this has been much less of an issue in the last two weeks since we've been back.
- it appears to me that Jake's keratosis pilaris is almost completely gone from his arms. He has had this as long as I can remember and there have never been periods when it is gone
- he seems to need more stimulation, sometimes in unproductive ways like chewing or licking the furniture, but he is less likely to get "wound up" by some of his repetitive actions. Previously if he was spinning a ball (he likes to watch things spin) he would get hyped up and start chewing on his tongue, drooling, and making moaning noises. Now he needs more of these things (watching TV, spinning balls, having us read books, having us sing to him) but it doesn't seem to get him out of control
- perhaps a slight improvement in communication, pointing to things that he wants or needs help with. No change in expressive language which is our biggest area of concern.
As far as we know, this is the first time a hit from a drug repurposing screen in fly was tested in a kiddo. No need for a mice model. No need for a cell model even.

Based on Jake’s multi-week promising exploratory single-patient study and in coordination with a clinical care team, Jake was taken off the drug (washout period) at the same time as two other MAN1B1-CDG families went on drug. This past weekend was the 4-week mark.
Excited to share more soon!