PGAP3 yeast drug multipurposing updates
We just completed our largest screen to date: the 12,500+ compound ReFRAME library from Scripps. We'll retest the top 1% of hit compounds in dose responses next, followed by the compound ID reveal.
Just before Christmas, we completed the first chunk of the ReFRAME library, as described here. Last week, all 52 384-well ReFRAME library plates were screened at the SMDC at UCSF in three consecutive batches. We don’t yet know the identities of the hit compounds — the unveiling happens after we collect dose-response data — but we can examine the dataset as a whole in the meantime.
Collaborators
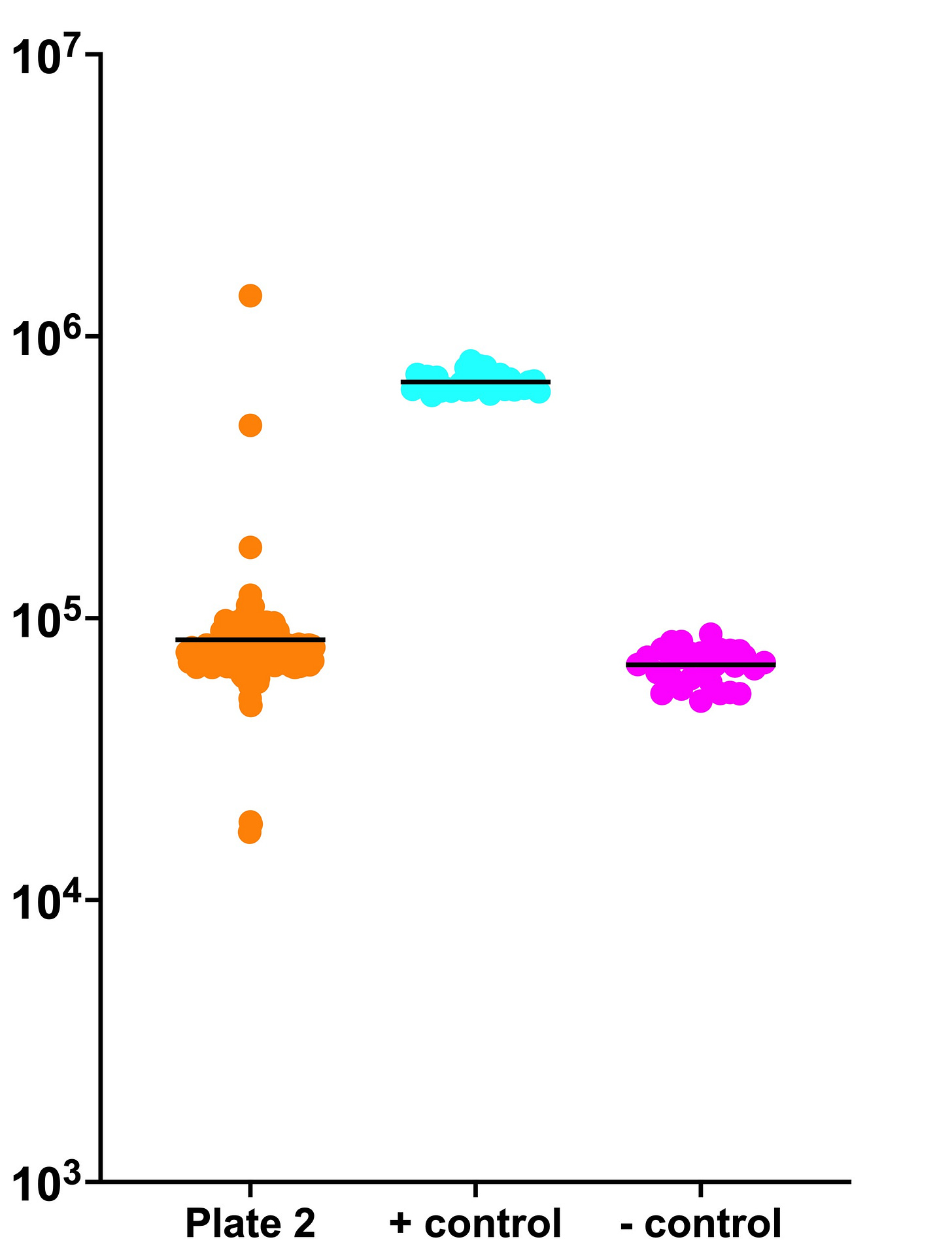
Below is a summary plot of unnormalized luminescence units (y-axis) of fifty-one 384-well plates — approximately 14,000 compounds along the x-axis — that comprise the ReFRAME collection. As described previously, we screened test compounds for growth rescue of per1∆/per1∆ homozygous diploid mutant yeast at 37˚C. Black lines indicate plate averages.
Even without normalization, it’s clear there are six strong hits which equal or in two instances exceed growth of the positive control. All of the orange circles below 10^4 can be considered enhancers, though some of these will be non-specifically fungistatic/fungitoxic, i.e., inhibit any yeast cell strain at the dose tested.
Plate 28 was dropped during the screening process and is in the queue to be redone later this month. Once we have the complete ReFRAME dataset we will share normalized Z scores of the complete dataset.
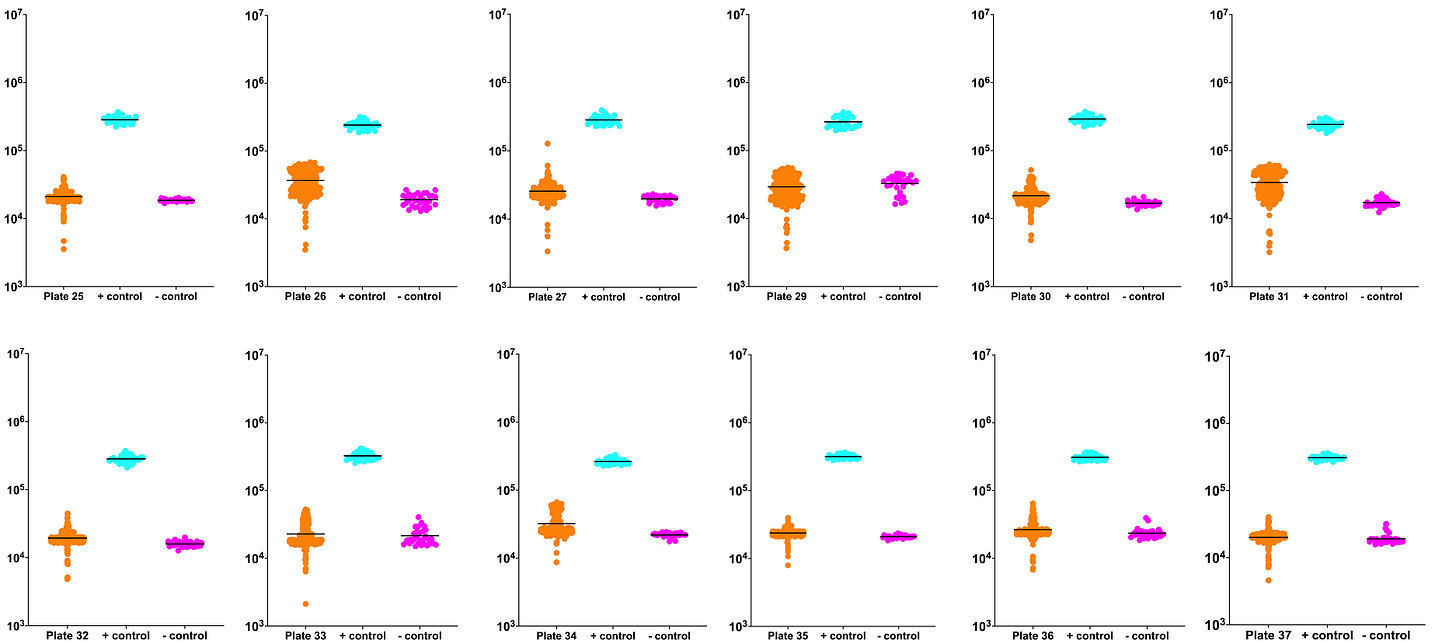
In the meantime, you can visually inspect the unnormalized ReFRAME dataset, plate by plate. The orange data points are test compounds. The cyan data points are the positive control (wildtype strain). The magenta data points are the negative control (untreated per1∆/per1∆ mutant).
A few of the plates show signs of edge or well effects which causes the orange data cloud to be dispersed. This happens when, for example, the left-half of the 384-well plate had higher luminescence raws than the right-half of the plate.
By the third and final batch of plates, position effects were resolved as shown by the plots for plates 50, 51 and 52. Normalization will take care of position effect and accentuate the weak-to-moderate hits.
The raw data will be normalized and spot-checked in the next few days. Then we submit hit compound well ID’s for up 1% of the library to Calibr @ Scripps Research, which in turn will ship back to us dose-response plates for a final round of hit testing. After we submit the dose-response data to the Calibr team, the compound names and structures will be revealed to us. We’re currently estimating that the big reveal will take place next month.