PGAP3 yeast drug repurposing updates
We completed the Pharmakon screen of 2,200 compounds which led to 2 antioxidants becoming part of Lucy's daily nutraceutical cocktail. We're 1/3 done with the ReFRAME library and we already have hits.
Last time when we provided a project update two months ago, we preliminarily analyzed the first of two rounds of drug repurposing screens commissioned by Moonshots for Unicorns. With the assistance of the Small Molecule Discovery Center located at UCSF, we screened the 2,200-compound Pharmakon library, which is an amalgam of two FDA-approved-drug libraries. Here we present an expanded analysis of the PGAP3 Pharmakon dataset and preview raw data from the ongoing second round of screening with the 12,000+ compound best-in-class drug repurposing library called ReFRAME.
Collaborators
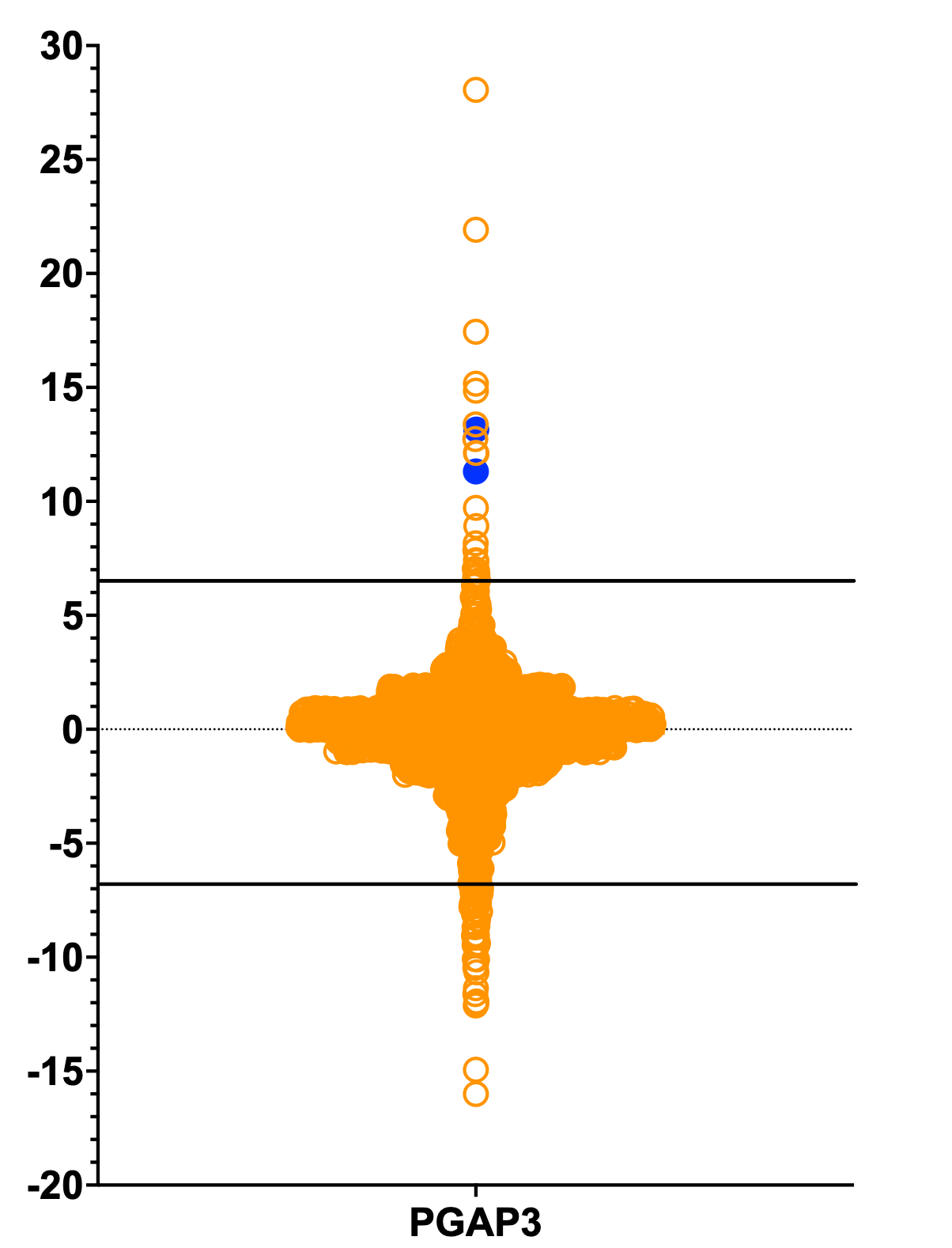
Below is a plot of Z-scores (y-axis) of the 2,200 compounds in the Pharmakon collection that were screened for rescue of growth of per1∆/per1∆ homozygous diploid mutant yeast at 37˚C, which we summarized in the inaugural post in this series. (Recall that the yeast gene version of PGAP3 is called PER1). The average Z-score value is -0.18. In line with the hit rates of previous yeast growth rescue screens, 26/2200 (1.18%) compounds have what may be considered a statistically significant growth rescue effect. A more stringent cutoff lowers the hit rate to below 1%.
Two hits (colored blue) are alpha lipoic acid (ALA) and folic acid, which are commonly used antioxidant supplements used in so called mito cocktails as treatments for mitochondrial diseases. In many ways PGAP3 presents more as a mitochondrial disease with decompensations triggered by immune system activation from infection or injury, which is not a clinical presentation typically seen in a classical congenital disorder of glycosylation. The results of disease modeling experiments on Lucy’s astrocytes and neurons sync up with the mechanisms of action of the Pharmakon PGAP3 yeast hits. Lucy has been taking ALA and folic acid since we transmitted the PGAP3 Pharmakon screening results at the beginning of October.
Last month we shared results from the SURF1 deficiency yeast drug repurposing project in partnership with Cure Mito Foundation. We were curious to see how the PGAP3 yeast hits performed in the SURF1 screen, and vice versa. With the exception of one compound with an antiviral mechanism of action, all of the PGAP3 yeast hits have no rescue effect in the SURF1 screen (left plot). In other words, there is no overlap between the PGAP3 hits and the SURF1 hits. In fact, most SURF1 yeast hits skew toward a growth-inhibitory effect in the PGAP3 screen. In other words, more compounds than expected reside in the upper left quadrant: SHY1 deficiency growth rescue but PGAP3 deficiency growth inhibition.
While we don’t see any overlap between PGAP3 hit and MEPAN hits, i.e., the upper right quadrant is empty, we also don’t observe the global leftward shift of the data points in a comparison between the PGAP3 screen and the MEPAN screen (right plot). The lower left quadrant — growth inhibition in both PGAP3 and MEPAN screens — shows a stronger correlation, which is expected as most compounds that live in this quadrant are across-the-board cytotoxic at the dose used in the primary screen. We don’t have enough data at this time to draw any biologically or clinically meaningful conclusions, but that will likely change when our drug repurposing data corpus grows large enough to have explanatory power and make testable predictions.
Here’s a plot of the raw data (y-axis is in luminescence units) for the first 6 plates of the ReFRAME library, roughly on par with the number of compounds in the Pharmakon collection. Excitingly, there are four hits that float above the data cloud. We’ll learn the identity of those hit compounds at the end of the screen. 20 ReFRAME plates were successfully screened this week, and we will complete the remaining 32 plates next month right after the winter break.
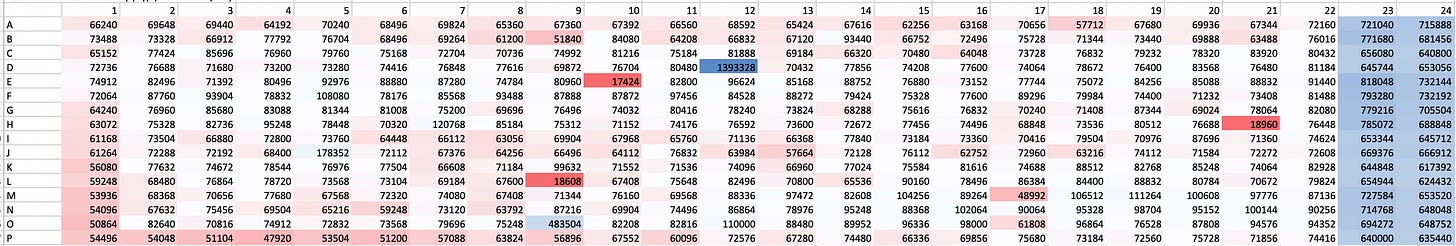
Here’s a heat map of the plate that contains the top growth rescuing hit compound. Notice that the level of growth supported by that top hit (dark blue, well D12) exceeds the level of growth achieved by the positive control wild-type yeast strain by a factor of two (light blue, rightmost two columns).
We can’t wait to wrap up the ReFRAME PGAP3 screen by mid-January. And then start to advance other yeast projects to the ReFRAME screening stage.