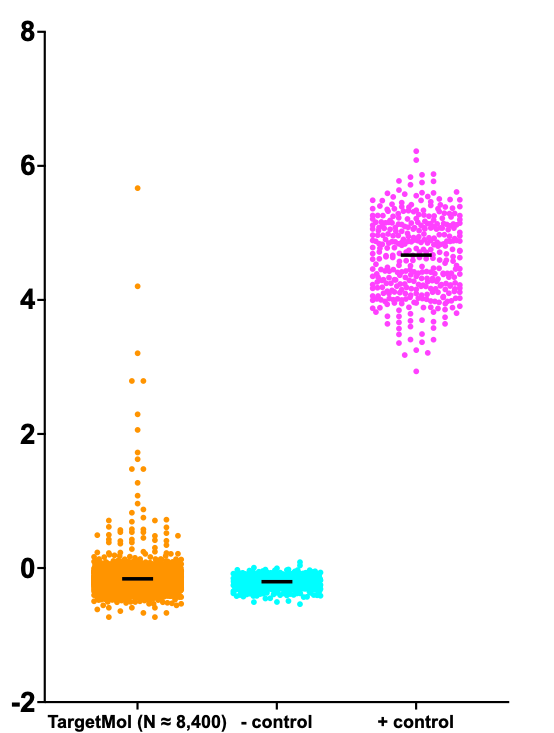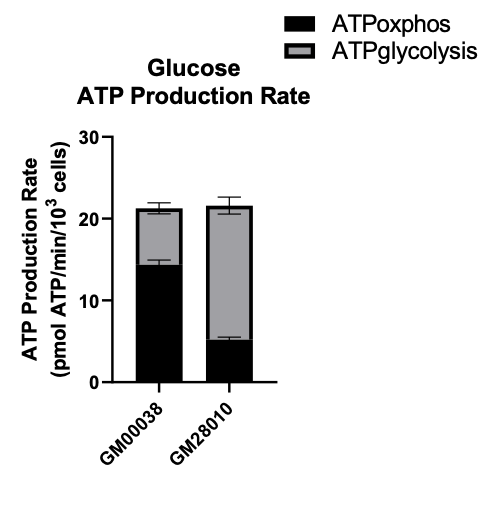SURF1 yeast hits validated in patient fibroblasts
Cure Mito Foundation partnered with Perlara on yeast-powered drug repurposing screens for SURF1 Leigh Syndrome. We found three compounds that increase mitochondrial respiration in SURF1 patient cells.
In collaboration with
Last year Cure Mito Foundation made a bet on Perlara and yeast as a drug repurposing platform. The bet has paid off preclinically, so far. For the first time, a rational mitochondrial cocktail is in sight if we can demonstrate therapeutic effect and elucidate rescue mechanisms in human cells.
Yet it will take more than just a silver bullet monotherapy to defeat a disease as pernicious and complex as Leigh Syndrome. Combination therapies will one day be the norm for inborn errors of metabolism, and likely most genetic diseases.
In analogy to Vertex’s Trikafta, a transformative novel triple combination therapy for cystic fibrosis, we envision combining three different (in this case repurposed) molecules for SURF1 deficiency, with each compound contributing an additive — or possibly synergistic — rescue mechanism. That’s a very ambitious goal still on the horizon, but we’re rocketing forward. We have no choice.
A year ago we completed our first SURF1 drug repurposing screen using the 2,250-compound Pharmakon library at the Small Molecule Discovery Center at UCSF. Six months ago, we completed a second SURF1 screen using the much larger 13,500-compound ReFRAME library, which had been made available to researchers by Scripps Research. Results of the Pharmakon and ReFRAME screens are summarized here.
In the spirit of completeness, we embarked on a third SURF1 screen using the 8,400-compound TargetMol library, which is now our first-line option for yeast-powered drug repurposing. Here’s a Z score summary plot of the SURF1 TargetMol dataset. The TargetMol data are in line with the Pharmakon and ReFRAME results.
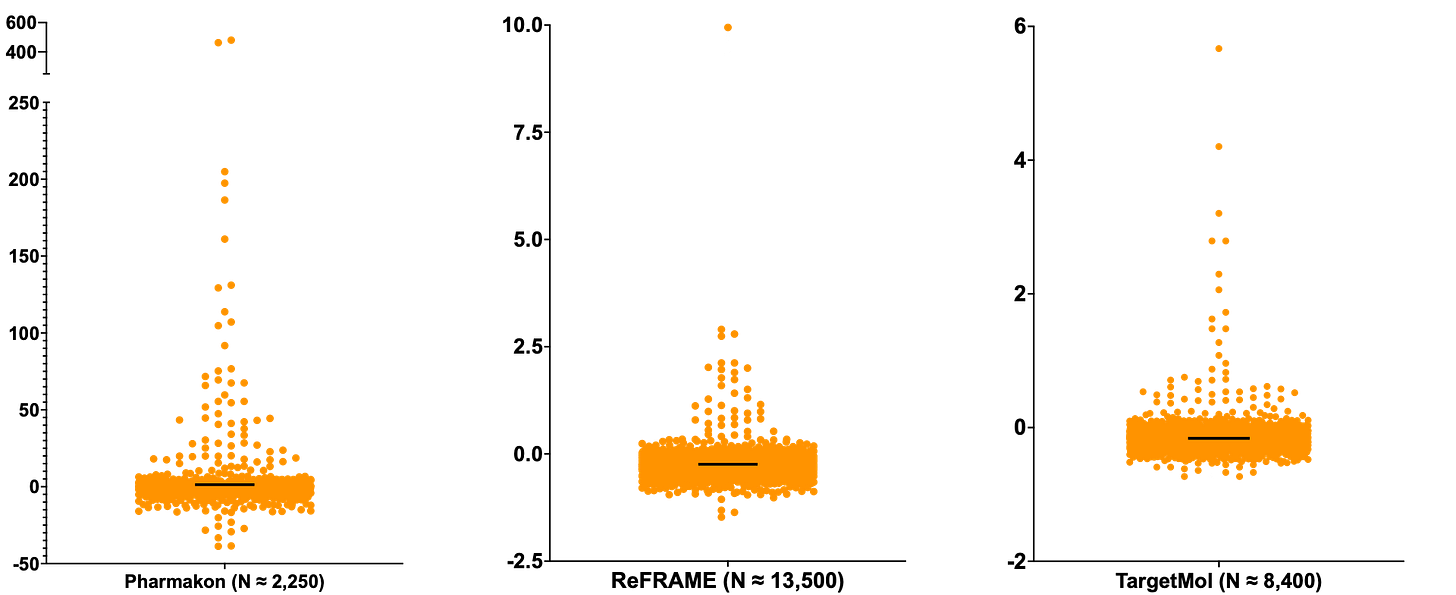
A head-to-head comparison of the Z score distributions across the three different drug repurposing libraries totaling 24,150 compounds is shown below. The shape of the Z score distributions is similar in all three screens. The Pharmakon screen had the lowest experimental variance and therefore much higher Z scores. Not too surprising given that there are only seven 384-well plates in the entire Pharmakon library, which also meant the screen was performed in a single run. In contrast, there are 52 plates in the ReFRAME library and 27 plates in the TargetMol library. Those libraries were screened in batches at different times.
Each library yielded dozens of rescuers. Notably, each screen resulted in a unique top rescuer. Consistent across all the libraries, there are only a handful number of sensitizers, especially relative to the number of rescuers. We don’t know what the excess of rescuers relative to sensitizers means other than that there may be multiple SURF1 rescue (or bypass) mechanisms available to activate pharmacologically.
After applying filters like pediatric approval status, oral bioavailability, and blood-brain barrier penetrance, we down-selected 12 yeast hits from the three libraries and reordered fresh powder stocks for mitochondrial stress tests in SURF1 patient fibroblasts. Linsey Stiles is the scientist at the UCLA Mitochondria and Metabolism core facility who performed the experiments summarized below.
The Seahorse instrument measures oxygen consumption rate (OCR) and also distinguishes between ATP production by glycolysis versus mitochondrial respiration (aka oxidative phosphorylation/OxPhos). As shown in the schematic below, cells are exposed to a series of stress tests that target the start, middle and end of the electron transport chain.
We tested a SURF1 patient fibroblast line (GM28010) and an age-matched control fibroblast line (GM00038). First, we established baselines measurements under standard growth condition — glucose as the carbon source — in the absence of compound treatment.
As shown in the figure below, ATP production from mitochondrial respiration (oxphos) is reduced by ~67% in the SURF1-deficient patient fibroblast line. There’s a compensatory increase in ATP production by glycolysis. What we’re looking for is a compound that increases ATP production by mitochondrial respiration, returning the disease profile back to normal.
Technically, four out of 12 yeast hits increased ATP production by mitochondrial respiration in a representative SURF1 patient fibroblast line. The fourth yeast hit increased basal respiration and ATP-linked respiration in galactose-cultured cells; however, this compound showed some toxicity in glucose-cultured cells with a dose-dependent decrease in all respiratory parameters measured.
Therefore, we considered three out of 12 yeast hits as validated. That translates to a 25% bioactivity conversion rate from a yeast cell to a human cell. Hereafter the three yeast hits will be referred to as Compounds 1-3.
Treatment with Compound 1 for 48 hours resulted in 12-15% boosts in ATP production by mitochondrial respiration and maximal respiration in both glucose-cultured and galactose-cultured cells. There is also concomitant decrease in ATP production through glycolysis.
Treatment with Compound 2 for 48 hours did not result increase maximal respiration in either glucose or galactose conditions. However, there was an increase in ATP production through both mitochondrial respiration and glycolysis.
Treatment with Compound 3 for 48 hours did not result in changes to maximal respiration in either glucose or galactose conditions, but there was an increase in ATP production through mitochondrial respiration at the 10 µM concentration.
Let’s look closer at maximal respiration in response to treatment with Compound 1 in galactose media versus glucose media.
Seven of the twelve yeast hits appear not to be active in fibroblasts.
Could it be because they’re yeast-specific hits, i.e., target a mechanism that is not evolutionarily conserved? Or do we need to treat with compound longer, or in different SURF1 patient fibroblast lines from genetically unrelated individuals? Or perhaps fibroblasts are the wrong cell type to study and we need to be interrogating neurons or muscle or other high-energy-demand cells. We’re pursuing the neuron option with Prof Alessandro Prigione’s lab in Germany.
Interestingly, two of the twelve yeast hits selectively affected ATP production by glycolysis. One of the hits decreases the fraction of ATP produced by glycolysis, while the other hit increases the fraction of ATP produced by glycolysis.
Altogether, half of the 12 SURF1 yeast hits are bioactive in SURF1 patient fibroblasts, a remarkable testament to the predictive power of Perlara’s yeast-powered drug repurposing platform. For any remaining skeptics who doubt the therapeutic relevance of yeast, remember that statistic: 50% conversion rate.
Next steps are to repeat the Seahorse assay on the 12 yeast hits in a genetically unrelated SURF1 patient line, and test combinations of Compounds 1-3. Onward!