Yeast-powered drug repurposing screens for SURF1 Leigh Syndrome are done 💊♻️ ✅
On behalf of Cure Mito Foundation, we completed high-throughput drug screens and dose response studies in yeast. Next up: validating hits in SURF1 patient fibroblasts and Complex IV yeast avatars.
In collaboration with
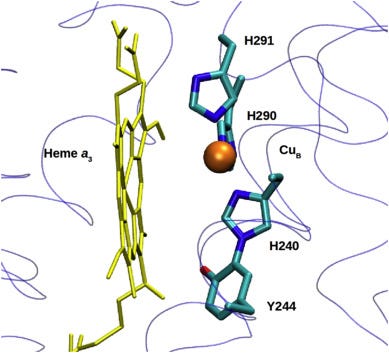
The featured image depicts Complex IV aka cytochrome c oxidase (COX) in all its macromolecular glory: 14 proteins huddled in the mitochondrial inner membrane, forming the reactor core of cellular metabolism where oxygen is reduced to water and the proton motive force is generated. If COX isn’t assembled correctly in a carefully choreographed sequence of protein folding and subunit binding steps, the result is a family of rare mitochondrial diseases called Leigh Syndrome.
Earlier this month, we wrapped up the drug screening stage of our yeast-enabled SURF1 Leigh Syndrome drug repurposing project. We’re about to begin the hit validation stage where we assess the effects of compounds on mitochondria function in SURF1 patient fibroblasts. On a parallel spur, we’ll assess COX enzyme activity in SURF1-deficient yeast in a background of strategically selected COX active site mutants.
After sweeping across a chemical space of 10,000+ unique compounds that includes FDA approved drugs, repositionable drugs that passed Phase 1 safety testing, generally recognized as safe nutriceuticals, and preclinical tool compounds, we identified 20-30 hits that rescue the growth of SURF1-deficient yeast cells grown on lactate challenge media, i.e., a nutrient broth that forces the yeast to use their mitochondria to grow.
It’s a stringent screen: OxPhos, or die. You might think the only way to fix the root cause of SURF1 deficiency is to replace the broken gene at the DNA level. The molecular details are still murky and were mostly worked out a decade ago in aerobic bacteria, but without the SURF1 protein chaperone the 14-protein Complex IV can’t assemble the active site, which involves insertion of heme followed by copper and the formation of an exotic post-translational modification called a histidine-tyrosine crosslink.
Without SURF1, cytochrome C oxidase can’t find alignment. Could a 300 molecular weight compound replace the function of a 300 amino acid protein like SURF1 by acting directly on the COX active site? Results so far suggest to us that some of the rescuer hits we’ve identified are acting as pharmacological fixes: small molecules that realign an active site that is out of whack.
And what if one of those rescuer hits is sitting on pharmacy shelves right now? Even if we couldn’t identify a small molecule that targets the root cause of SURF1 deficiency and restores COX activity, there could be Complex IV bypass pathways or other forms of compensation that allow a SURF1-deficient yeast cell to power oxidative phosphorylation.
SURF1 was our first drug repurposing project using an off-the-shelf whole-gene knockout avatar for a nuclear-encoded, mitochondrial-localized protein. There are dozens of genes like SURF1 that are compatible with the following three-stage plug-and-play process. What you might call yeast drug repurposing as service:
First stage is procuring MATa and MATalpha haploid mutants and the homozygous diploid mutant, and then comparing them to haploid and diploid wildtype controls on solid media in a low-throughput, ten-fold dilution series.
Second stage is confirming the growth defect in a 96-well plate liquid format using the Bac-TiterGlo (BTG) luminescence assay, which is more sensitive albeit costlier than a simple absorbance assay.
Stage three is confirming the growth defect is robust in 384-well plate liquid format. This is the final step of assay optimization, followed by a drug screen.
We aspire to complete hit validation studies in SURF1 patient fibroblasts over the next few months. The initial recap of the SURF1 Pharmakon screen gave us confidence in the yeast model but also reminded us how the first SURF1-deficient yeast mutant was characterized over two decades ago. The hot-of-the-presses ReFRAME summary confirmed that we were in for a treat.
Once we had the complete Pharmakon and ReFRAME datasets, we could prioritize rescuer hits in part by focusing on the overlap between the two screens. That’s the focus of this post. We observed such an overlap, at least in terms of pharmacological classes, in the PGAP3 drug repurposing project that we wrapped up two months ago.
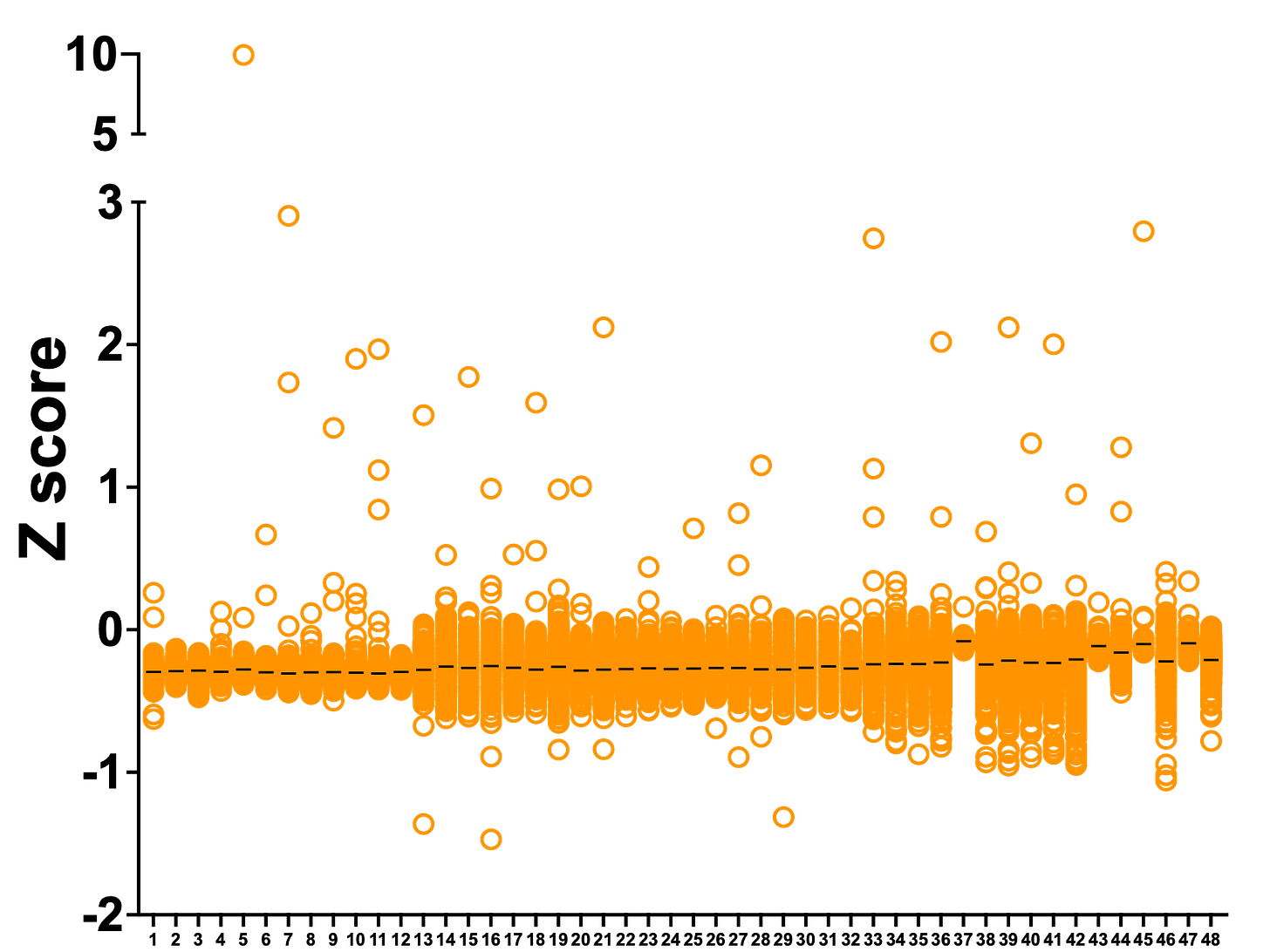
In PGAP3, one compound was a hit in both datasets. In SURF1, five compounds are hits in both datasets. Note that we removed 8 false positives (grey circles) from the ReFRAME dataset. Tantalizingly, the top ReFRAME hit with a Z score just under 10 is not present in the Pharmakon library.

The Z scores are much larger in the Pharmakon screen because the variance was smaller. The Pharmakon library is made up of seven 384-well plates that can be screened in one go. On the other hand, the ReFRAME library is 48 plates that were screened in three batches. Hence the larger variance and lower absolute Z scores. The results are no less meaningful, it’s merely a function of performing the screen in batches. (Moving forward, we’ll be working with the HTSF at Cal where we’ll be able to screen a 48-plate library in a single run).
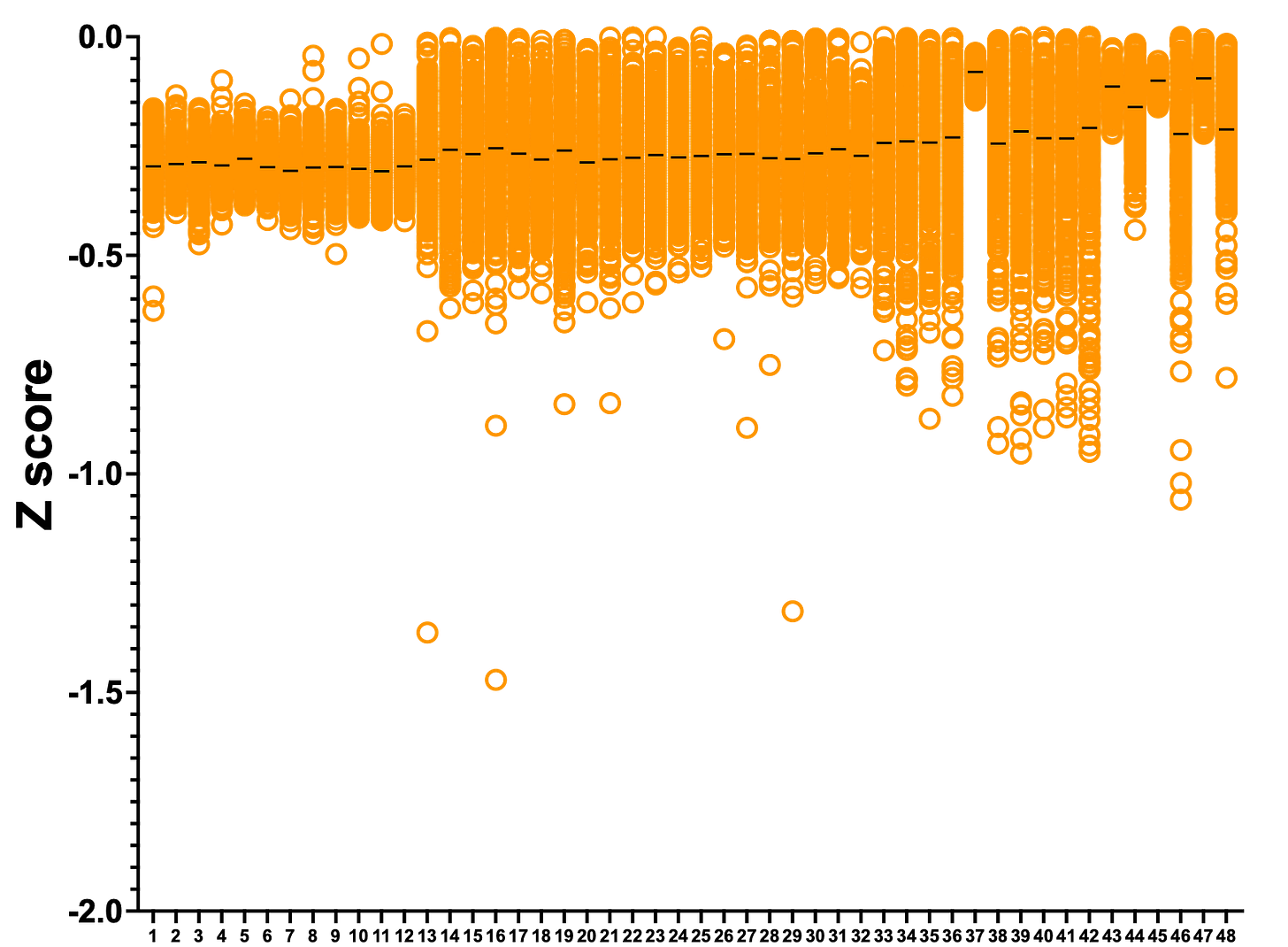
Removing the false positive outliers, we plotted the ReFRAME dataset where each column is one of the 48 plates that comprise the library. Lucky plate #5 has the top hit, which we’ll describe more below.
Rescuer hits are evenly distributed across the 48 library plates. Interestingly, there are only three strong sensitizing hits. One handwavy explanation: the shy1∆ knockout yeast avatar is not viable in lactose media, so it’s difficult to make it even more sick.
How does SURF1 compare to MECR, a gene that causes an ultra-rare mitochondrial disease called MEPAN? The pairwise comparisons below reveal a handful of common hits but mostly gene-specific hits. A few SURF1 rescuers are MEPAN sensitizers. MEPAN has more sensitizers overall.
Next, let’s compare SURF1 and PGAP3. SURF1 rescuers are skewed toward being PGAP3 sensitizers, while PGAP3 rescuers are inert in SURF1. We’re not certain of the biological significance but it points to the duality of rescue mechanisms.
Reassuringly, SURF1 rescuers are gene-specific. In other words, these are not promiscuous compounds that somehow rescue the growth of any sick yeast mutant.
114 primary screening positives were selected, including manual selection of plate winners and losers. In total, 77 presumptive rescuers plus 37 presumptive sensitizers were requested from Calibr in dose response plates with technical replicates.
Below is the top hit from the primary screen, which is also the most active rescuer in a dose response experiment. This compound rescued SURF1-deficient yeast growth to twice the level achieved by the positive control, a wildtype yeast with intact SURF1 grown on lactate.
Two of the five compounds that are hits in both Pharmakon and ReFRAME screens are shown below. These fall into the intermediate rescuers camp, though it should be noted that this chemical series has sub-microlar potency.
We saw a few biphasic hits, too. For example:
Let’s blow out the y-axis to exaggerate the sensitizers. Although they won’t be therapeutics, sensitizers can teach us about how rescuers work.
The top three sensitizers are actually two compounds, as one of them appeared in the library twice (serving as added layer of internal replication). Curiously, both compounds are dyes. Neither are sensitizers in the PGAP3 screen, so this appears to be a SURF1-specific result.
Trypan blue is vital stain. It’s used to tell if a cell is dead or not under the microscope. Gentian violet has been in use as a vital stain in the lab, as a textile dye and as an early 20th century anti-infective medicine. Why they are acting as sensitizers is a head scratcher at the moment.
However, two other sensitizers stand out in a way that makes biological sense based on what we know about SURF1 function: the porphyrins hemin and stannsoporfin. In the absence of SURF1, there is accumulation of excess heme that is not incorporated into the COX active site. So addition of exogenous heme in the form of hemin would presumably push a SURF1-deficient yeast cell over the edge.
Stannsoporfin is described as heme oxygenase inhibitor. Heme oxygenase is the first enzyme in the heme breakdown pathway. So an inhibitor of heme oxygenase would result in an intracellular buildup of heme in a SURF1-deficient yeast cell that is already experiencing heme stress.
Based on characterization of bacterial SURF1, which is conserved with human SURF1 and yeast SURF1, lacking SURF1 is expected to cause a hypersensitivity to excess heme. Quoting one of the published SURF1 biochemical studies:
The interaction of Surf1 with heme a synthase and subunit I as well as its heme binding capacities suggest a reservoir function of Surf1 that controls the flux of heme a from its site of biosynthesis to its final target in oxidase subunit I.
That paper goes on to conclude:
The findings in yeast suggest a role for Surf1 as a chaperon that cotranslationally recruits de-novo synthesised helices of subunit I to keep them in an open conformation, thus enabling insertion of its heme cofactors.
Once we have data from hit validation studies, we’ll be ready to share more about the SURF1 rescuers. We want more evidence in support of the pharmacological fix model that certain old drugs are directly potentiating or activating Complex IV and thereby pharmacologically replacing the function of SURF1.
We hope to be able to share more at the upcoming UMDF Mito Medicines meeting next month. Onward!











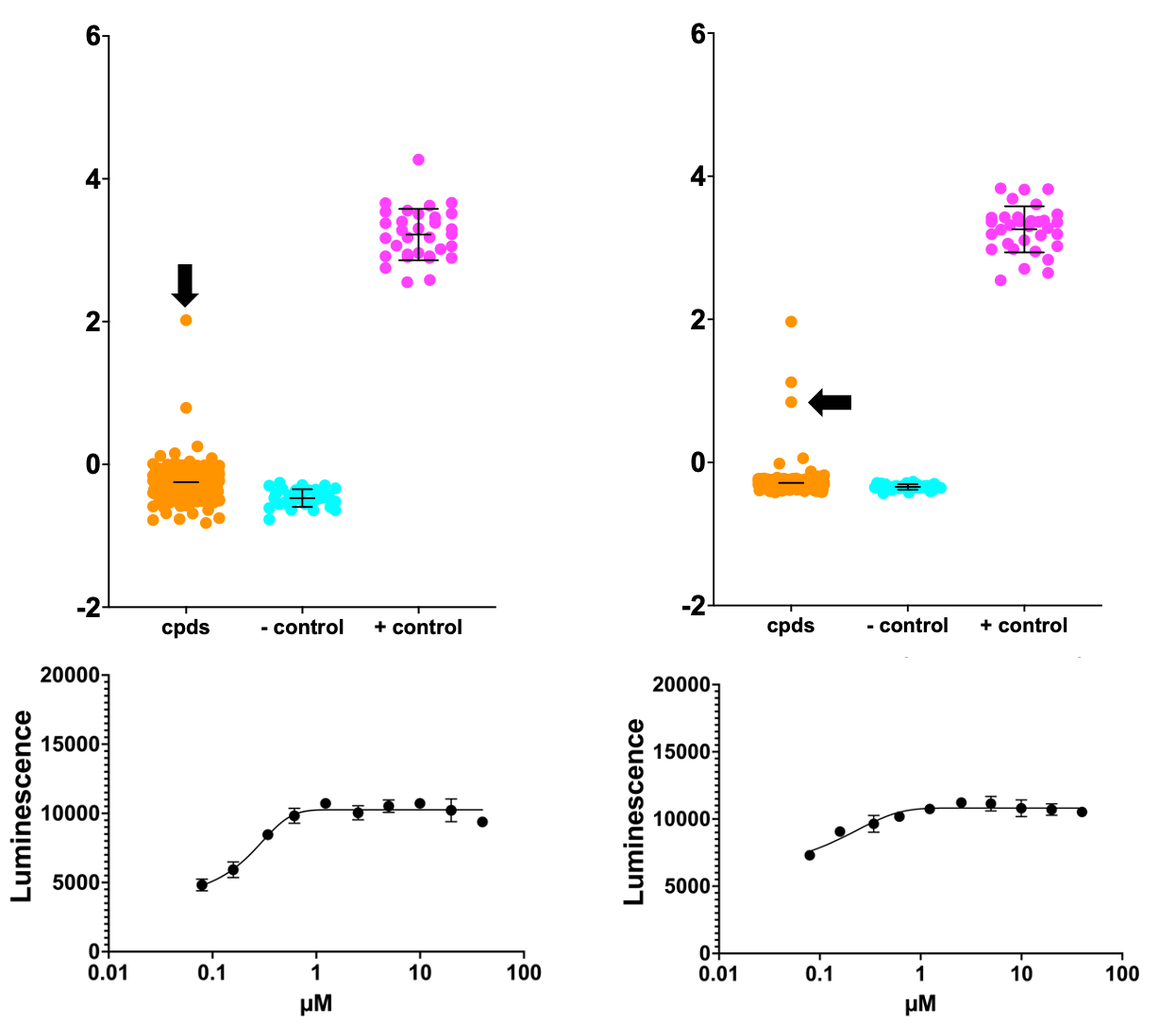





This is fascinating! Is this 10k+ chemical library available to license or is it a proprietary toolset?
Thanks again for these regular updates on your screening. I enjoy being kept up to date on the developments rather than all together in a final research paper. The sensitisers are an interesting set of compounds worthy of further research. Do you think the rescuers found in both the SURF1 and PGAP3 point to more general survival or growth mechanisms than gene specific rescue? Keep up this wonderful project