PGAP3 drug repurposing screens are done 💊♻️ ✅
On behalf of Moonshots for Unicorns, we completed the Pharmakon and ReFRAME drug screens using a yeast model of PGAP3 deficiency. We validated over 100 hits including both rescuers and sensitizers.
In collaboration with
We’re excited to announce the completion of primary drug screens and secondary hit validation studies for the PGAP3 yeast project sponsored by Moonshots for Unicorns.
Let’s recap the journey that began 10 months ago when we got introduced to Drs. Geri and Zachary Landman by Dr Maura Ruzhnikov. After initial conversations where we deliberated the pros and cons of working with a centralized lab in academia, we decided to conduct two novel experiments: one experiment in distributed biotech operations, and a parallel experiment in yeast-powered personalized medicine.
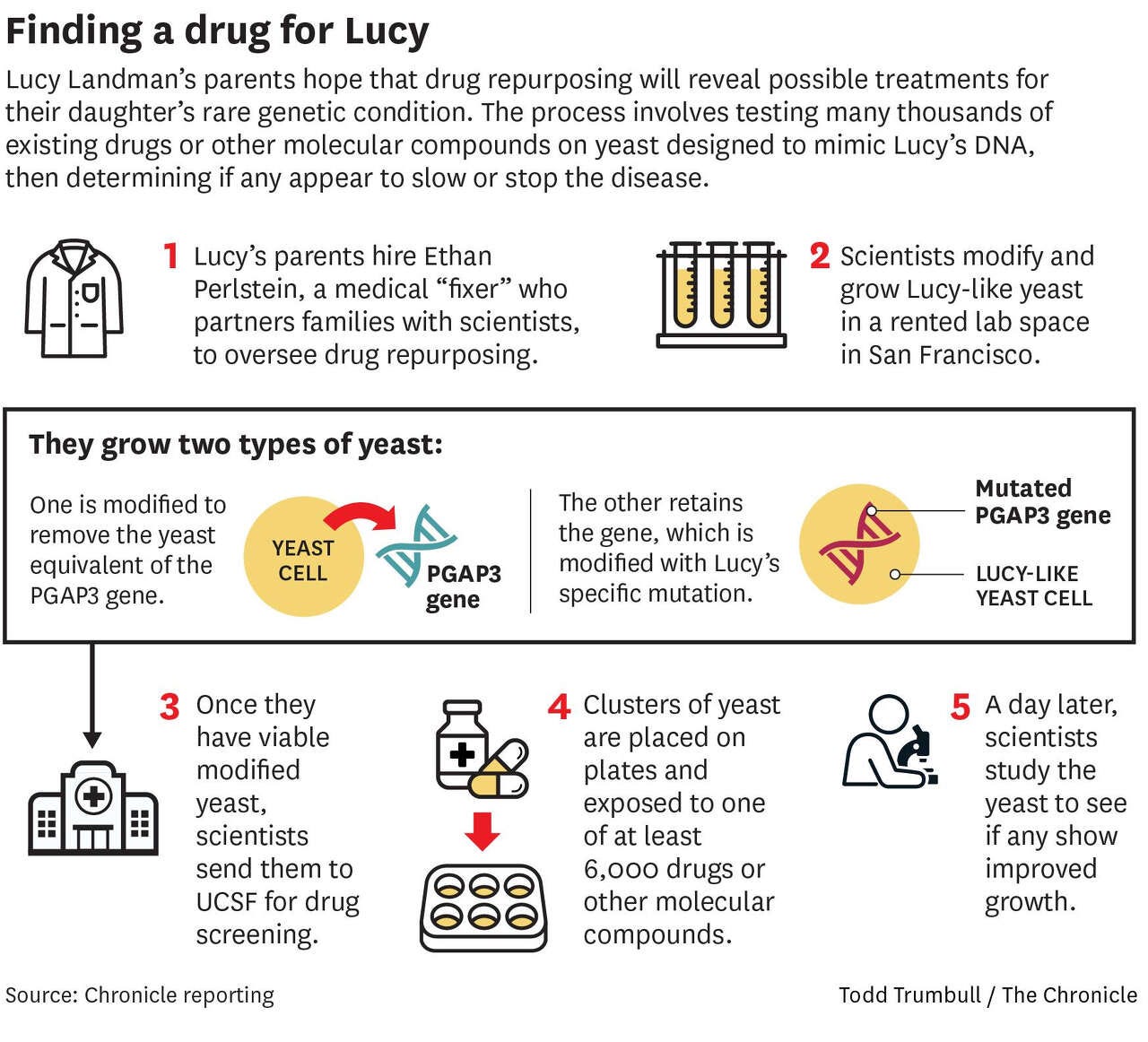
An infographic created for a story by San Francisco Chronicle health reporter Erin Allday summarized the plan on paper. In practice, we made a few tactical pivots along the way. For example, we doubled down on the PGAP3-knockout yeast avatar versus spending more resources on troubleshooting a yeast avatar with Lucy’s variants. Some steps took longer than expected because we had to run an extra round of optimization or screening. We tried to iterate and learn as fast as possible.
Part 1: May - August 2022
Last summer, we popped up a cure lab in the heart of San Francisco focused on drug repurposing screens using yeast models of inherited monogenic metabolic diseases. PGAP3 was our inaugural project.
We recruited three amazing Gen Z scientists from UCSF as the first experimentalist Perlara Cure Guides: Henry Ng, Dana Kennedy and Kelly Pan. Yeast team leader Dr. Mathura Thevandavakkam remotely managed the yeast lab, updating Gantt charts on Smartsheets and troubleshooting in real-time on Slack from her home in Michigan, three time zones away. Doctoral candidate Justin Donnelly, a graduate student in Professor Carolyn Bertozzi’s lab at Stanford, was responsible for client communications and data analysis. CDG Program Director Dr. Kristin Kantautas oversaw the project from the Netherlands.
We didn’t need a brand-new lab space or much more than basic microbiology lab infrastructure: orbital shaker, incubator, freezer, fridge, etc. We did all the yeast growth characterization and early drug screening assay optimization at the San Francisco popup lab. Then there were handoffs of yeast cultures to Dr. Jez Revalde at the Small Molecule Discovery Center at UCSF. Jez performed the actual drug screens, and then reported back the data to us.
In the month of August, while most people were enjoying their summer vacation, we sprinted to lay the groundwork for the pilot drug screen of the Pharmakon library.
Part 2: September - October 2022
After a 30-day sprint in August to ensure that the PGAP3-knockout yeast mutant strains behaved in the lab as expected, at the start of September we handed the baton to Jez at the SMDC to perform the final drug screening assay optimization experiments, followed by the Pharmakon screen itself. We settled on the diploid homozygous PGAP3-knockout mutant, which performed the best in head-to-head assessments with the MATa and MATα haploid PGAP3-knockout mutants.
On Friday September 30, the 7-plate Pharmakon library was successfully screened. The following Monday October 3, we got access to the data and emailed the results to Moonshots. What ensued was a whirlwind sequence of scientific firsts.
Immediately, we learned from the Landmans that Professor Kathrin Meyer’s lab at Nationwide Children’s Hospital in Ohio had independent evidence implicating iron-induced cellular toxicity in Lucy’s induced pluripotent stem cell derived astrocyte model. Lucy’s iPSC neural progenitor cells are dying of iron overload, Zach replied in the rapidly expanding October 3 email thread.
One of the top Pharmakon hits is a clinically approved iron chelator. We’d never seen seen convergence between a yeast avatar and a patient-derived neuronal cell model so quickly before!
Cooler still, one of the Pharmakon hits is an over-the-counter supplement available on Amazon. It’s thought to alleviate iron toxicity by a mechanism that involves cooper. Based on the striking iron toxicity convergence, the Landmans ordered up the compound and gave some to Lucy. At that time she was 15 months old and had stopped babbling at 8 months. At a Perlara happy hour meetup in the East Bay on Wednesday October 5, Zach told us that the morning after her first dose, Lucy woke up babbling again.
After a few weeks the Landmans stopped giving the Lucy the compound due to tolerability issues, but there were what we might call glimmers of efficacy.
Part 3: November 2022 - January 2023
After we analyzed the Pharmakon dataset and the Landmans acted on several other yeast hits that are known components of mitochondrial cocktails, such as folic acid and alpha lipoic acid, we were eager to screen a comprehensive compound collection to identify all possible PGAP3 bypass or compensatory rescue pathways.
Luckily, we learned about the opportunity to screen the ReFRAME collection maintained by the Scripps Institute. We applied for access to ReFRAME based on our success with the Pharmakon collection. Fortunately, we were accepted. In early December, Jez at the SMDC received fifty-two 384-well plates. Over the next four weeks, all 52 plates were screened. Unfortunately, one of the 52 plates was accidentally dropped and had to be redone, resulting in an unexpected delay of several weeks.
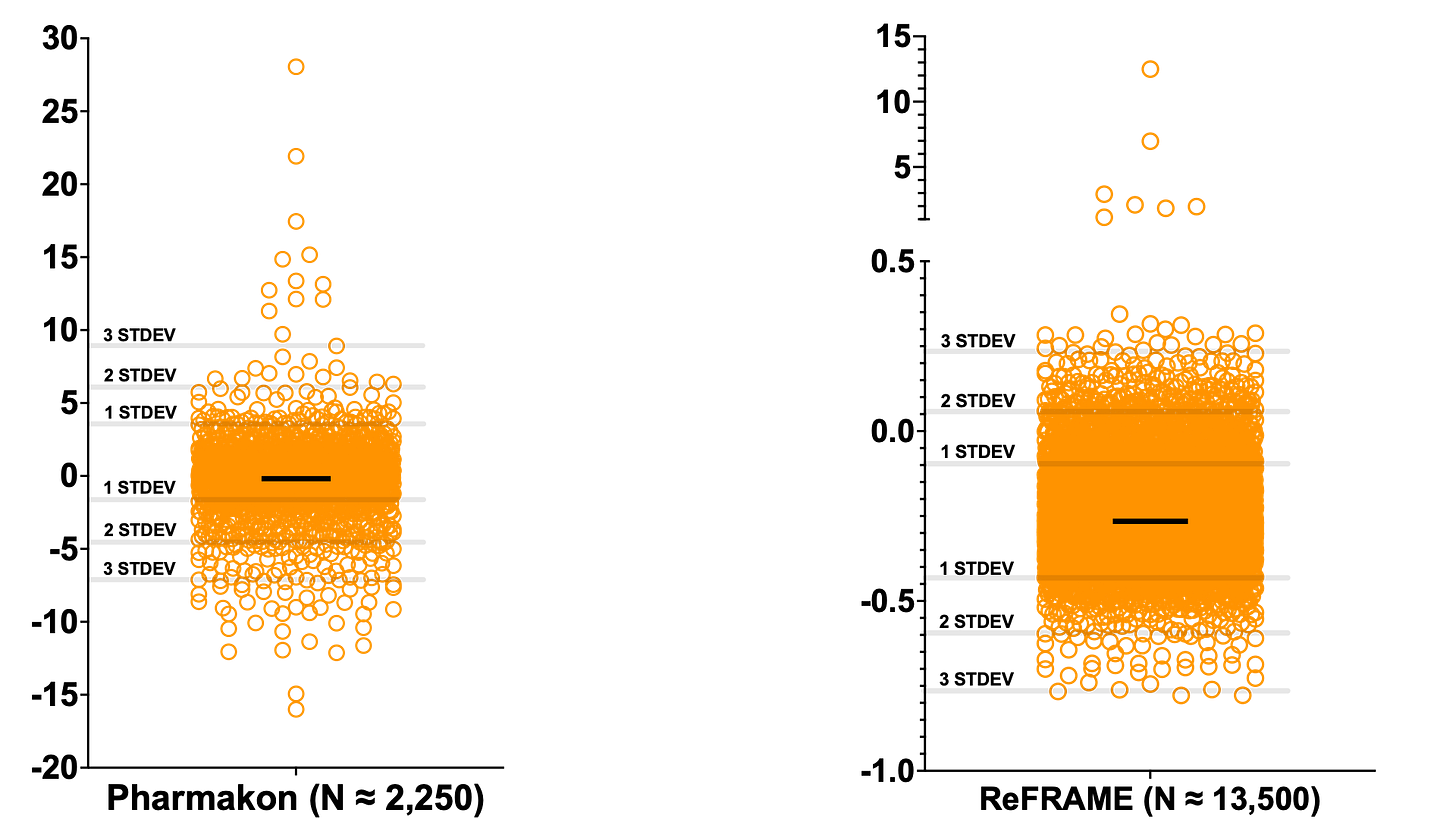
In spite of that one unexpected operational hiccup, we ended up with what we believe is a statistically robust, first-of-its-kind drug repurposing dataset for PGAP3 deficiency made possible by yeast. Plotted below are Z-score distributions. They look remarkably similar.
Part 4: February - March 2023
The lone straggler plate was re-screened in mid-February. We then requested dose-response plates for the top 1% of hits, approximately 100 unique compounds. We selected all hits with Z scores more than 2 standard deviations from the mean. We also cherrypicked plate winners that did not meet the two standard deviations cutoff.
Ten dose response plates arrived at the SMDC a month later in mid-March. On Wednesday March 22, wildtype and PGAP3-knockout yeast strains were transported a few miles across town from the SF popup lab to the SMDC. These yeast were then dispensed into the ten dose-response plates. The next day, we received a csv file with the raw data.
Three classes of hit compounds are indicated below. The rescuers are orange-filled circles. The sensitizers are red-filled circles. Co-mingling among the rescuers and the sensitizers, we observed a third class of hit compounds that have a biphasic dose response curve: growth inhibition at high doses, but growth rescue at low doses. Those so-called biphasics are the blue-filled circles.
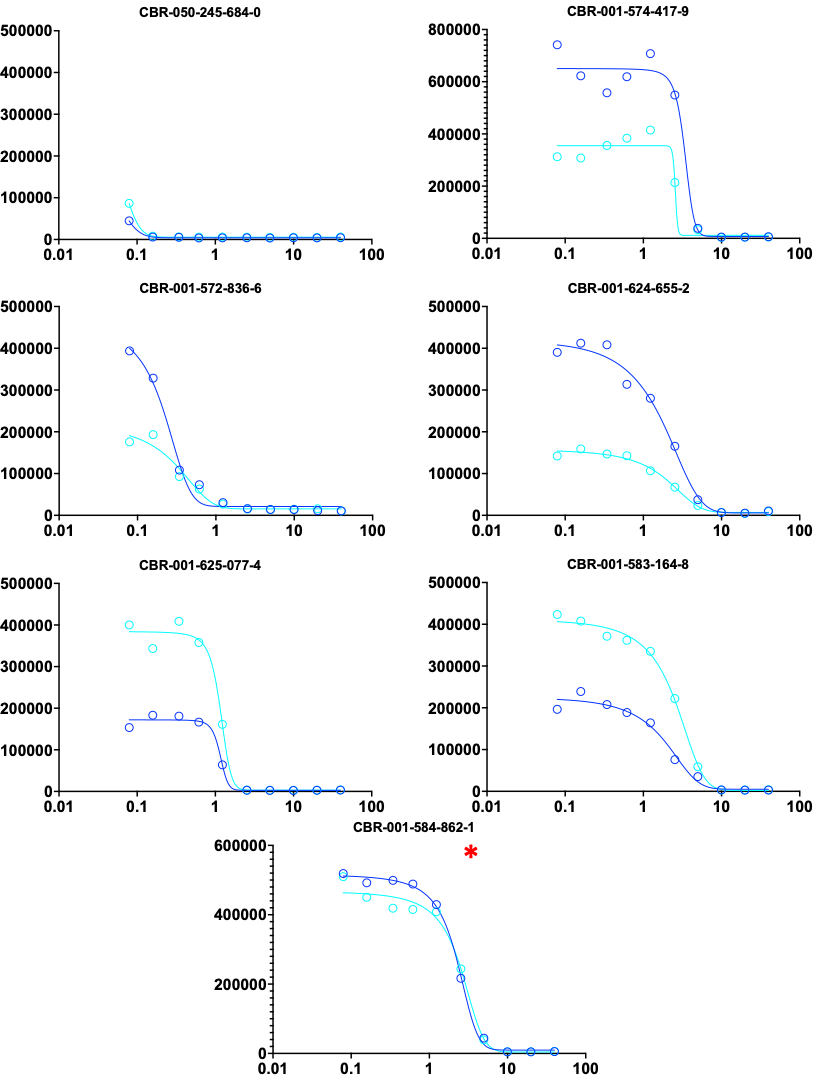
Here are the dose-response curves for the top-6 rescuers (one of the rescuers was present in the library twice). Interestingly, the rescuer with the highest Z score in the primary screen was not the most potent rescuer in the dose-response hit validation studies.
Below, the four rescuers on the left have a nice sigmoidal curve and excellent agreement between both technical replicates (blue circles versus cyan circles). Curiously, the three rescuers on the right do not show evidence of plateauing.
Here are the four biphasic compounds that rescue at lower doses but sensitize at higher doses. Their mechanism of action is very likely to be distinct from the classic rescuers.
Finally, here are the top-7 sensitizers, including the compound marked by a red asterisk that has a neutral Z-score, i.e., is in the cloud with the rescuers and biphasics. These sensitizer curves are the mirror image of the rescuer curves above.
How many different rescue mechanisms are at play?
Are the sensitizers targeting the same pathways as the rescuers but with opposite effect?
What are the biphasics doing?
We’ll attempt to answer those questions in a followup analysis soon.