SURF1 ReFRAME recap
On behalf of Cure Mito Foundation, last week we completed a comprehensive 13,500-compound drug repurposing screen using a Leigh Syndrome yeast avatar of SURF1 deficiency.
In collaboration with
Since the last SURF1 Leigh Syndrome drug repurposing project update a month ago, we’re pleased to announce that we completed the ReFRAME library screen of ~13,500 compounds!
This is our second ReFRAME library screen. (The first was for the PGAP3 drug repurposing project). Earlier this week, we submitted a list of 114 compounds to Calibr/Scripps for hit validation studies. We hope to receive the dose-response plates in the next few weeks. In parallel, we’re still testing the SURF1 Pharmakon rescue hits on the SURF1-deficient yeast in a cytochrome c oxidase (COX) activity assay.
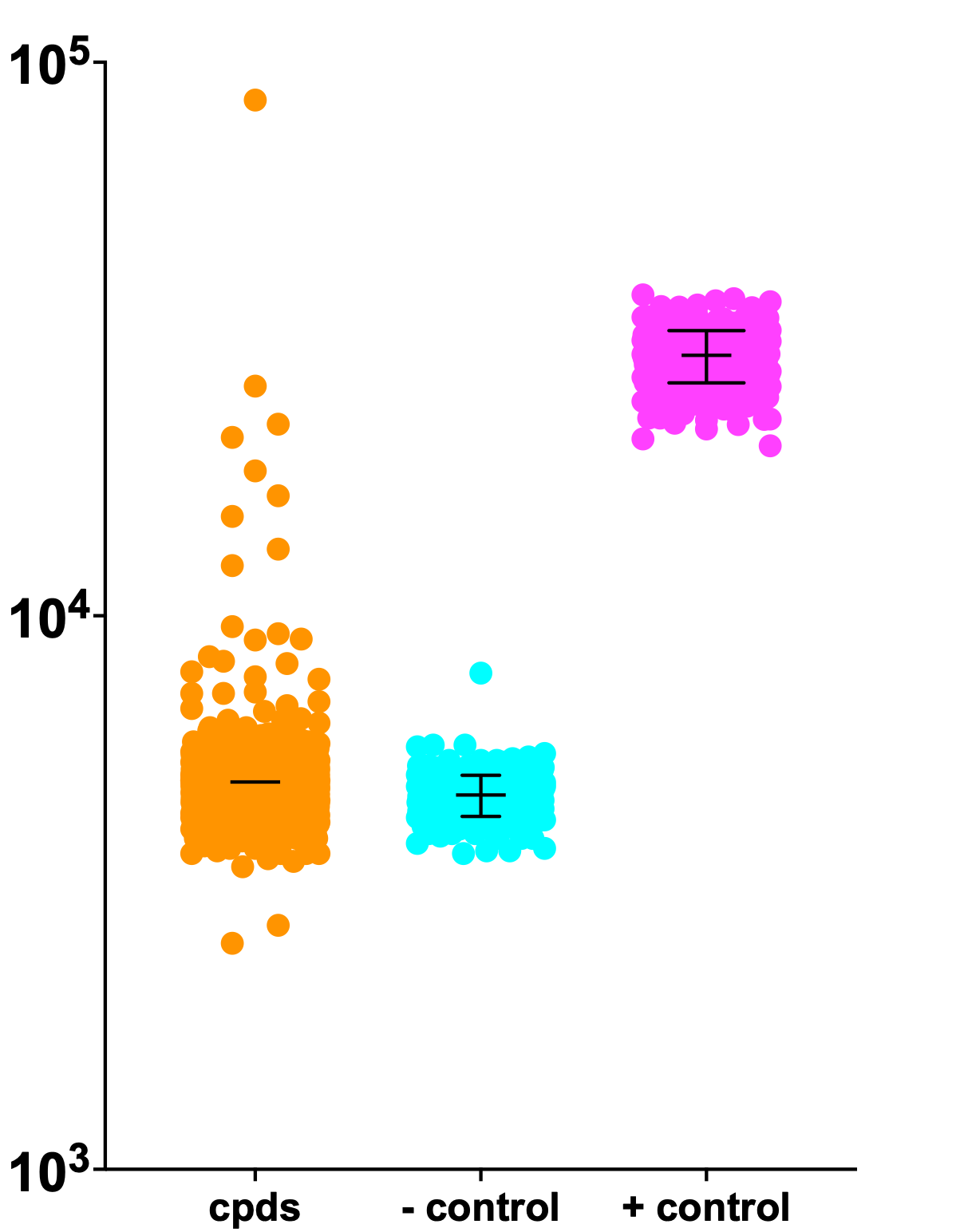
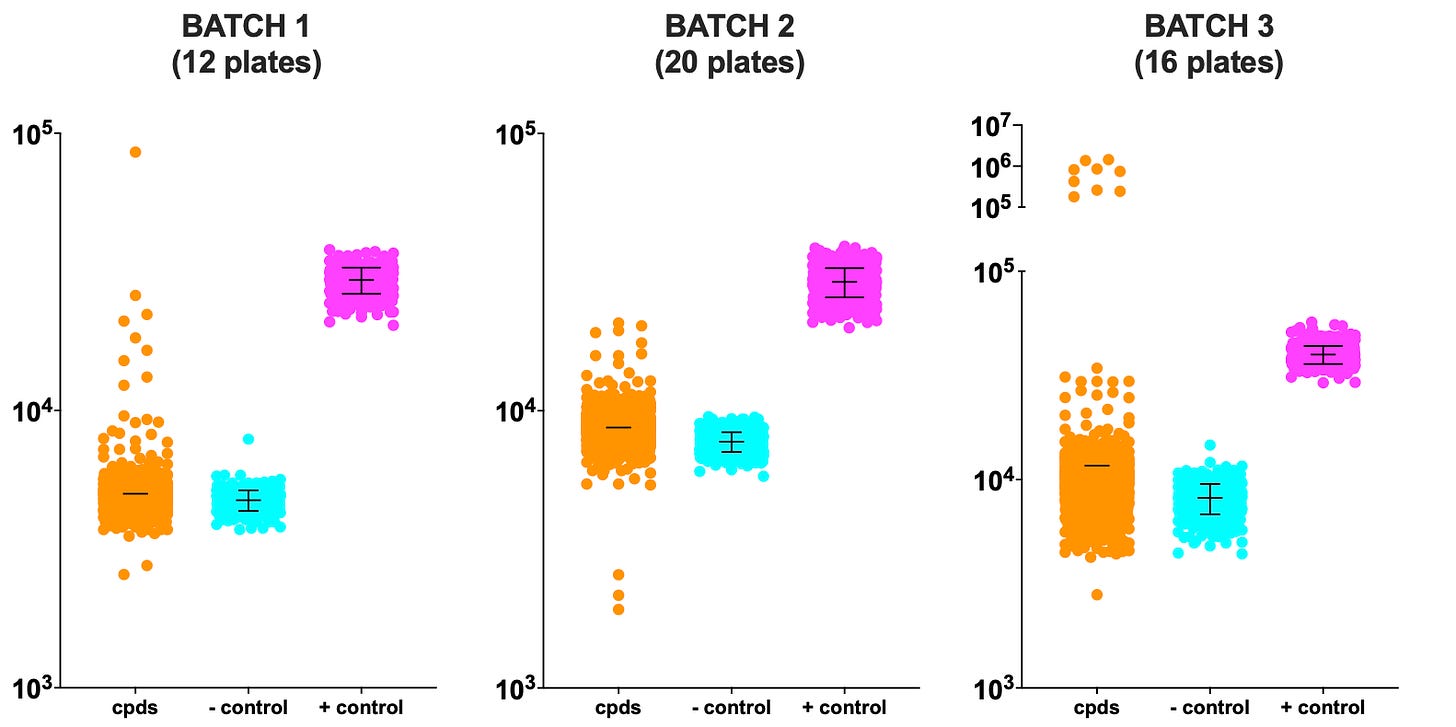
In the meantime, let’s kick the tires on the SURF1 ReFRAME dataset. Below is the unnormalized (raw) data. The screen was performed in three batches. In all batches, there is clear separation between the negative control and the positive control. Batch 3 has the most variance. Hits with luminescence values greater than 10^5 (100,000) are restricted to the third batch. Suspiciously, we observed similar “high flying” hits in the PGAP3 ReFRAME screen and as we’ll explain below, these hits are indeed too good to be true.
That said, as we saw in the SURF1 Pharmakon screen, Batch 1 includes a rescue hit that exceeds the positive control values, and several rescuing hits that achieve growth comparable to the positive control. Batches 2 and 3 also have hits that overlap with the positive control values.
Here's what the normalized ReFRAME dataset looks like when we plot Z scores (y-axis) instead of raw luminescence counts:
Next we visualized the unnormalized dataset from the perspective of each of the 48 ReFRAME library plates. While there is a discernible batch effect, we are still able to call hits by eye. Batch 3 has more weak sensitizing hits (sensitizers) and the per-plate variance is increased relative to the first two batches but is still in an acceptable range. Overall, the rescuing hits (rescuers) outnumber the sensitizers, which is consistent with the ratio we observed in the Pharmakon SURF1 screen.
Based on our recent experience with PGAP3, we knew to be on the lookout for ATP analogs as false positives. Recall that our standard yeast growth assay is based on luminescence generated by the luciferase enzyme, which is fueled by ATP. Indeed, we confirmed that each of the nine high-flying SURF1 hits are the same false positives in the PGAP3 screen.
When we remove the nine false positives, here’s the plot of Z scores for ~13,5000 compounds in the ReFRAME library:
And here’s the Z score data plotted per plate with the false positives removed:
Stay tuned for the next update on the SURF1 Leigh Syndrome yeast drug repurposing project!









This is a really interesting study, I look forward to finding out about the hits you find. For me SURF1 is a really interesting protein and maybe these hits can help to understand the function of this protein.